Analyzing Epistemic and Aleatoric Uncertainty for Drusen Segmentation in Optical Coherence Tomography Images
Paper and Code
Feb 08, 2021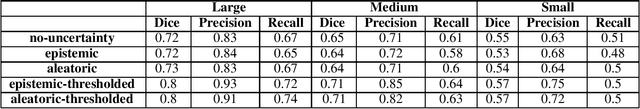
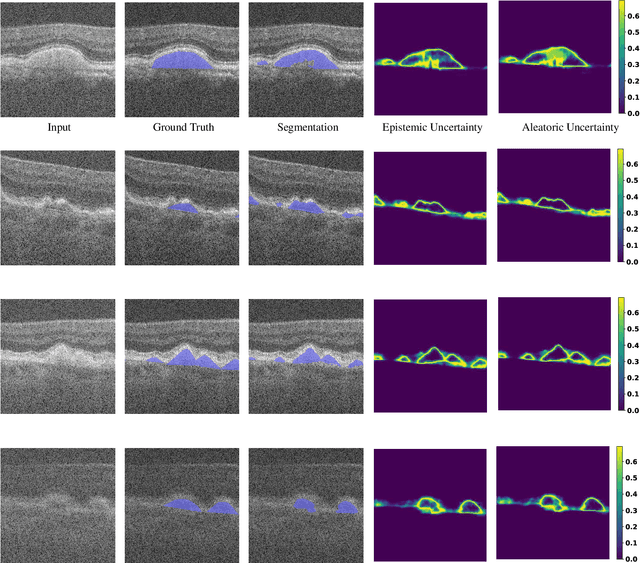
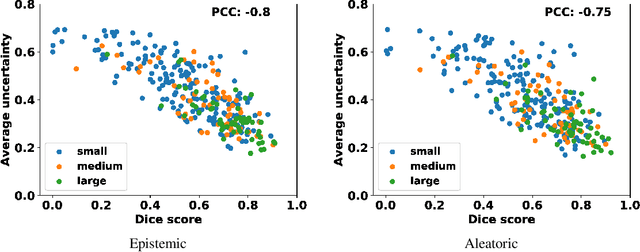
Age-related macular degeneration (AMD) is one of the leading causes of permanent vision loss in people aged over 60 years. Accurate segmentation of biomarkers such as drusen that points to the early stages of AMD is crucial in preventing further vision impairment. However, segmenting drusen is extremely challenging due to their varied sizes and appearances, low contrast and noise resemblance. Most existing literature, therefore, have focused on size estimation of drusen using classification, leaving the challenge of accurate segmentation less tackled. Additionally, obtaining the pixel-wise annotations is extremely costly and such labels can often be noisy, suffering from inter-observer and intra-observer variability. Quantification of uncertainty associated with segmentation tasks offers principled measures to inspect the segmentation output. Realizing its utility in identifying erroneous segmentation and the potential applications in clinical decision making, here we develop a U-Net based drusen segmentation model and quantify the segmentation uncertainty. We investigate epistemic and aleatoric uncertainty capturing model confidence and data uncertainty respectively. We present segmentation results and show how uncertainty can help formulate robust evaluation strategies. We visually inspect the pixel-wise uncertainty and segmentation results on test images. We finally analyze the correlation between segmentation uncertainty and accuracy. Our results demonstrate the utility of leveraging uncertainties in developing and explaining segmentation models for medical image analysis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge