Stephen Bach
BigBIO: A Framework for Data-Centric Biomedical Natural Language Processing
Jun 30, 2022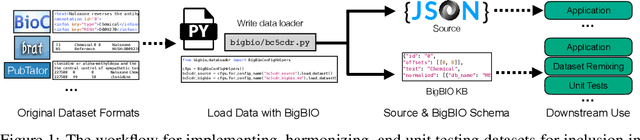
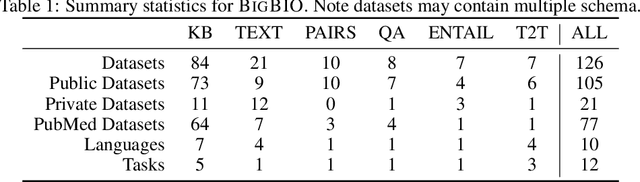
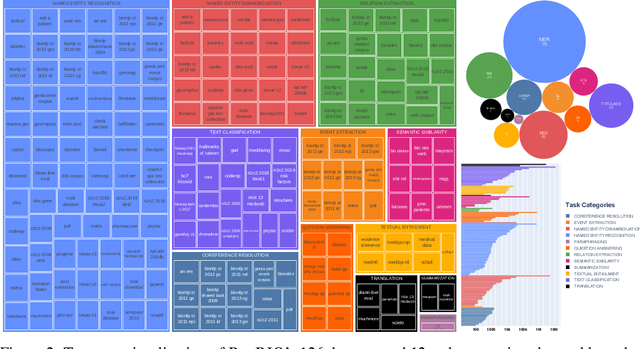
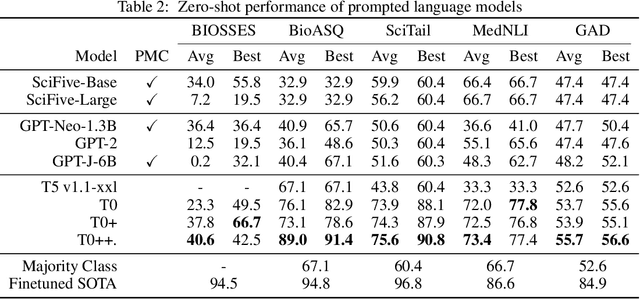
Abstract:Training and evaluating language models increasingly requires the construction of meta-datasets --diverse collections of curated data with clear provenance. Natural language prompting has recently lead to improved zero-shot generalization by transforming existing, supervised datasets into a diversity of novel pretraining tasks, highlighting the benefits of meta-dataset curation. While successful in general-domain text, translating these data-centric approaches to biomedical language modeling remains challenging, as labeled biomedical datasets are significantly underrepresented in popular data hubs. To address this challenge, we introduce BigBIO a community library of 126+ biomedical NLP datasets, currently covering 12 task categories and 10+ languages. BigBIO facilitates reproducible meta-dataset curation via programmatic access to datasets and their metadata, and is compatible with current platforms for prompt engineering and end-to-end few/zero shot language model evaluation. We discuss our process for task schema harmonization, data auditing, contribution guidelines, and outline two illustrative use cases: zero-shot evaluation of biomedical prompts and large-scale, multi-task learning. BigBIO is an ongoing community effort and is available at https://github.com/bigscience-workshop/biomedical
Hinge-loss Markov Random Fields: Convex Inference for Structured Prediction
Sep 26, 2013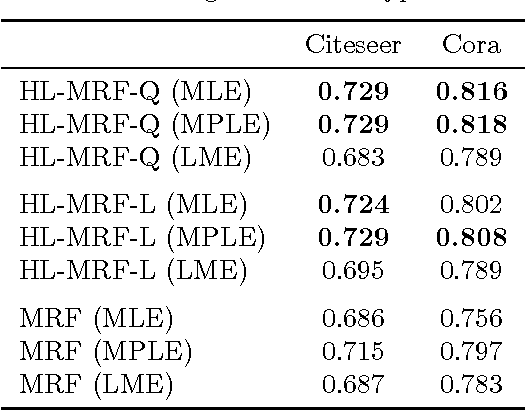

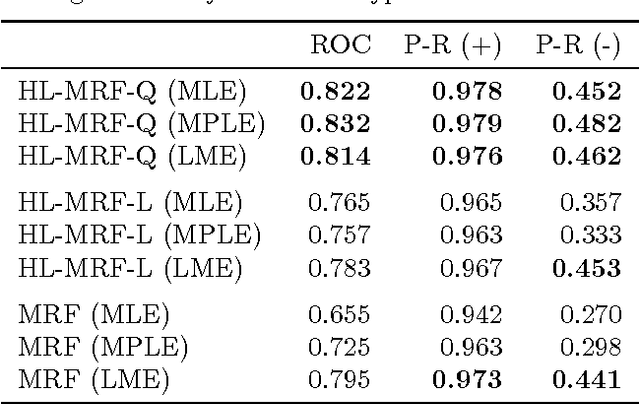
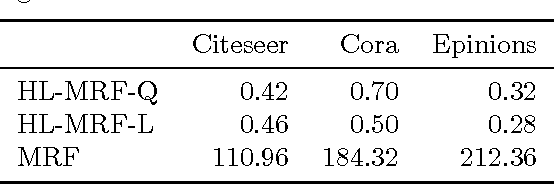
Abstract:Graphical models for structured domains are powerful tools, but the computational complexities of combinatorial prediction spaces can force restrictions on models, or require approximate inference in order to be tractable. Instead of working in a combinatorial space, we use hinge-loss Markov random fields (HL-MRFs), an expressive class of graphical models with log-concave density functions over continuous variables, which can represent confidences in discrete predictions. This paper demonstrates that HL-MRFs are general tools for fast and accurate structured prediction. We introduce the first inference algorithm that is both scalable and applicable to the full class of HL-MRFs, and show how to train HL-MRFs with several learning algorithms. Our experiments show that HL-MRFs match or surpass the predictive performance of state-of-the-art methods, including discrete models, in four application domains.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge