Stefan Kramer
Deep Generative Models of Evolution: SNP-level Population Adaptation by Genomic Linkage Incorporation
Jul 28, 2025Abstract:The investigation of allele frequency trajectories in populations evolving under controlled environmental pressures has become a popular approach to study evolutionary processes on the molecular level. Statistical models based on well-defined evolutionary concepts can be used to validate different hypotheses about empirical observations. Despite their popularity, classic statistical models like the Wright-Fisher model suffer from simplified assumptions such as the independence of selected loci along a chromosome and uncertainty about the parameters. Deep generative neural networks offer a powerful alternative known for the integration of multivariate dependencies and noise reduction. Due to their high data demands and challenging interpretability they have, so far, not been widely considered in the area of population genomics. To address the challenges in the area of Evolve and Resequencing experiments (E&R) based on pooled sequencing (Pool-Seq) data, we introduce a deep generative neural network that aims to model a concept of evolution based on empirical observations over time. The proposed model estimates the distribution of allele frequency trajectories by embedding the observations from single nucleotide polymorphisms (SNPs) with information from neighboring loci. Evaluation on simulated E&R experiments demonstrates the model's ability to capture the distribution of allele frequency trajectories and illustrates the representational power of deep generative models on the example of linkage disequilibrium (LD) estimation. Inspecting the internally learned representations enables estimating pairwise LD, which is typically inaccessible in Pool-Seq data. Our model provides competitive LD estimation in Pool-Seq data high degree of LD when compared to existing methods.
Enhancing Symbolic Machine Learning by Subsymbolic Representations
Jun 17, 2025Abstract:The goal of neuro-symbolic AI is to integrate symbolic and subsymbolic AI approaches, to overcome the limitations of either. Prominent systems include Logic Tensor Networks (LTN) or DeepProbLog, which offer neural predicates and end-to-end learning. The versatility of systems like LTNs and DeepProbLog, however, makes them less efficient in simpler settings, for instance, for discriminative machine learning, in particular in domains with many constants. Therefore, we follow a different approach: We propose to enhance symbolic machine learning schemes by giving them access to neural embeddings. In the present paper, we show this for TILDE and embeddings of constants used by TILDE in similarity predicates. The approach can be fine-tuned by further refining the embeddings depending on the symbolic theory. In experiments in three real-world domain, we show that this simple, yet effective, approach outperforms all other baseline methods in terms of the F1 score. The approach could be useful beyond this setting: Enhancing symbolic learners in this way could be extended to similarities between instances (effectively working like kernels within a logical language), for analogical reasoning, or for propositionalization.
Focus, Merge, Rank: Improved Question Answering Based on Semi-structured Knowledge Bases
May 14, 2025Abstract:In many real-world settings, machine learning models and interactive systems have access to both structured knowledge, e.g., knowledge graphs or tables, and unstructured content, e.g., natural language documents. However, most rely on either. Semi-Structured Knowledge Bases (SKBs) bridge this gap by linking unstructured content to nodes within structured data, thereby enabling new strategies for knowledge access and use. In this work, we present FocusedRetriever, a modular SKB-based framework for multi-hop question answering. It integrates components (VSS-based entity search, LLM-based generation of Cypher queries and pairwise re-ranking) in a way that enables it to outperform state-of-the-art methods across all three STaRK benchmark test sets, covering diverse domains and multiple performance metrics. The average first-hit rate exceeds that of the second-best method by 25.7%. FocusedRetriever leverages (1) the capacity of Large Language Models (LLMs) to extract relational facts and entity attributes from unstructured text, (2) node set joins to filter answer candidates based on these extracted triplets and constraints, (3) vector similarity search to retrieve and rank relevant unstructured content, and (4) the contextual capabilities of LLMs to finally rank the top-k answers. For generality, we only incorporate base LLMs in FocusedRetriever in our evaluation. However, our analysis of intermediate results highlights several opportunities for further upgrades including finetuning. The source code is publicly available at https://github.com/kramerlab/FocusedRetriever .
Human Guided Learning of Transparent Regression Models
Feb 21, 2025Abstract:We present a human-in-the-loop (HIL) approach to permutation regression, the novel task of predicting a continuous value for a given ordering of items. The model is a gradient boosted regression model that incorporates simple human-understandable constraints of the form x < y, i.e. item x has to be before item y, as binary features. The approach, HuGuR (Human Guided Regression), lets a human explore the search space of such transparent regression models. Interacting with HuGuR, users can add, remove, and refine order constraints interactively, while the coefficients are calculated on the fly. We evaluate HuGuR in a user study and compare the performance of user-built models with multiple baselines on 9 data sets. The results show that the user-built models outperform the compared methods on small data sets and in general perform on par with the other methods, while being in principle understandable for humans. On larger datasets from the same domain, machine-induced models begin to outperform the user-built models. Further work will study the trust users have in models when constructed by themselves and how the scheme can be transferred to other pattern domains, such as strings, sequences, trees, or graphs.
Integrating Inverse and Forward Modeling for Sparse Temporal Data from Sensor Networks
Feb 19, 2025Abstract:We present CavePerception, a framework for the analysis of sparse data from sensor networks that incorporates elements of inverse modeling and forward modeling. By integrating machine learning with physical modeling in a hypotheses space, we aim to improve the interpretability of sparse, noisy, and potentially incomplete sensor data. The framework assumes data from a two-dimensional sensor network laid out in a graph structure that detects certain objects, with certain motion patterns. Examples of such sensors are magnetometers. Given knowledge about the objects and the way they act on the sensors, one can develop a data generator that produces data from simulated motions of the objects across the sensor field. The framework uses the simulated data to infer object behaviors across the sensor network. The approach is experimentally tested on real-world data, where magnetometers are used on an airport to detect and identify aircraft motions. Experiments demonstrate the value of integrating inverse and forward modeling, enabling intelligent systems to better understand and predict complex, sensor-driven events.
Soft Hoeffding Tree: A Transparent and Differentiable Model on Data Streams
Nov 07, 2024
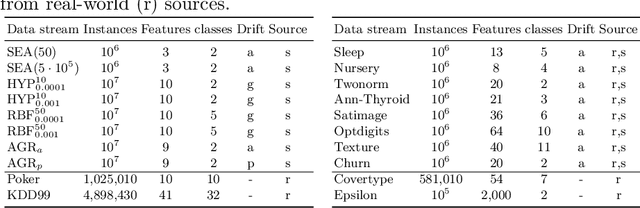


Abstract:We propose soft Hoeffding trees (SoHoT) as a new differentiable and transparent model for possibly infinite and changing data streams. Stream mining algorithms such as Hoeffding trees grow based on the incoming data stream, but they currently lack the adaptability of end-to-end deep learning systems. End-to-end learning can be desirable if a feature representation is learned by a neural network and used in a tree, or if the outputs of trees are further processed in a deep learning model or workflow. Different from Hoeffding trees, soft trees can be integrated into such systems due to their differentiability, but are neither transparent nor explainable. Our novel model combines the extensibility and transparency of Hoeffding trees with the differentiability of soft trees. We introduce a new gating function to regulate the balance between univariate and multivariate splits in the tree. Experiments are performed on 20 data streams, comparing SoHoT to standard Hoeffding trees, Hoeffding trees with limited complexity, and soft trees applying a sparse activation function for sample routing. The results show that soft Hoeffding trees outperform Hoeffding trees in estimating class probabilities and, at the same time, maintain transparency compared to soft trees, with relatively small losses in terms of AUROC and cross-entropy. We also demonstrate how to trade off transparency against performance using a hyperparameter, obtaining univariate splits at one end of the spectrum and multivariate splits at the other.
Harnessing the Power of Semi-Structured Knowledge and LLMs with Triplet-Based Prefiltering for Question Answering
Sep 01, 2024Abstract:Large Language Models (LLMs) frequently lack domain-specific knowledge and even fine-tuned models tend to hallucinate. Hence, more reliable models that can include external knowledge are needed. We present a pipeline, 4StepFocus, and specifically a preprocessing step, that can substantially improve the answers of LLMs. This is achieved by providing guided access to external knowledge making use of the model's ability to capture relational context and conduct rudimentary reasoning by themselves. The method narrows down potentially correct answers by triplets-based searches in a semi-structured knowledge base in a direct, traceable fashion, before switching to latent representations for ranking those candidates based on unstructured data. This distinguishes it from related methods that are purely based on latent representations. 4StepFocus consists of the steps: 1) Triplet generation for extraction of relational data by an LLM, 2) substitution of variables in those triplets to narrow down answer candidates employing a knowledge graph, 3) sorting remaining candidates with a vector similarity search involving associated non-structured data, 4) reranking the best candidates by the LLM with background data provided. Experiments on a medical, a product recommendation, and an academic paper search test set demonstrate that this approach is indeed a powerful augmentation. It not only adds relevant traceable background information from information retrieval, but also improves performance considerably in comparison to state-of-the-art methods. This paper presents a novel, largely unexplored direction and therefore provides a wide range of future work opportunities. Used source code is available at https://github.com/kramerlab/4StepFocus.
10 Years of Fair Representations: Challenges and Opportunities
Jul 04, 2024Abstract:Fair Representation Learning (FRL) is a broad set of techniques, mostly based on neural networks, that seeks to learn new representations of data in which sensitive or undesired information has been removed. Methodologically, FRL was pioneered by Richard Zemel et al. about ten years ago. The basic concepts, objectives and evaluation strategies for FRL methodologies remain unchanged to this day. In this paper, we look back at the first ten years of FRL by i) revisiting its theoretical standing in light of recent work in deep learning theory that shows the hardness of removing information in neural network representations and ii) presenting the results of a massive experimentation (225.000 model fits and 110.000 AutoML fits) we conducted with the objective of improving on the common evaluation scenario for FRL. More specifically, we use automated machine learning (AutoML) to adversarially "mine" sensitive information from supposedly fair representations. Our theoretical and experimental analysis suggests that deterministic, unquantized FRL methodologies have serious issues in removing sensitive information, which is especially troubling as they might seem "fair" at first glance.
Amplifying Exploration in Monte-Carlo Tree Search by Focusing on the Unknown
Feb 13, 2024



Abstract:Monte-Carlo tree search (MCTS) is an effective anytime algorithm with a vast amount of applications. It strategically allocates computational resources to focus on promising segments of the search tree, making it a very attractive search algorithm in large search spaces. However, it often expends its limited resources on reevaluating previously explored regions when they remain the most promising path. Our proposed methodology, denoted as AmEx-MCTS, solves this problem by introducing a novel MCTS formulation. Central to AmEx-MCTS is the decoupling of value updates, visit count updates, and the selected path during the tree search, thereby enabling the exclusion of already explored subtrees or leaves. This segregation preserves the utility of visit counts for both exploration-exploitation balancing and quality metrics within MCTS. The resultant augmentation facilitates in a considerably broader search using identical computational resources, preserving the essential characteristics of MCTS. The expanded coverage not only yields more precise estimations but also proves instrumental in larger and more complex problems. Our empirical evaluation demonstrates the superior performance of AmEx-MCTS, surpassing classical MCTS and related approaches by a substantial margin.
Peer Learning: Learning Complex Policies in Groups from Scratch via Action Recommendations
Dec 15, 2023Abstract:Peer learning is a novel high-level reinforcement learning framework for agents learning in groups. While standard reinforcement learning trains an individual agent in trial-and-error fashion, all on its own, peer learning addresses a related setting in which a group of agents, i.e., peers, learns to master a task simultaneously together from scratch. Peers are allowed to communicate only about their own states and actions recommended by others: "What would you do in my situation?". Our motivation is to study the learning behavior of these agents. We formalize the teacher selection process in the action advice setting as a multi-armed bandit problem and therefore highlight the need for exploration. Eventually, we analyze the learning behavior of the peers and observe their ability to rank the agents' performance within the study group and understand which agents give reliable advice. Further, we compare peer learning with single agent learning and a state-of-the-art action advice baseline. We show that peer learning is able to outperform single-agent learning and the baseline in several challenging discrete and continuous OpenAI Gym domains. Doing so, we also show that within such a framework complex policies from action recommendations beyond discrete action spaces can evolve.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge