Songhan Ge
ImplicitCell: Resolution Cell Modeling of Joint Implicit Volume Reconstruction and Pose Refinement in Freehand 3D Ultrasound
Mar 09, 2025Abstract:Freehand 3D ultrasound enables volumetric imaging by tracking a conventional ultrasound probe during freehand scanning, offering enriched spatial information that improves clinical diagnosis. However, the quality of reconstructed volumes is often compromised by tracking system noise and irregular probe movements, leading to artifacts in the final reconstruction. To address these challenges, we propose ImplicitCell, a novel framework that integrates Implicit Neural Representation (INR) with an ultrasound resolution cell model for joint optimization of volume reconstruction and pose refinement. Three distinct datasets are used for comprehensive validation, including phantom, common carotid artery, and carotid atherosclerosis. Experimental results demonstrate that ImplicitCell significantly reduces reconstruction artifacts and improves volume quality compared to existing methods, particularly in challenging scenarios with noisy tracking data. These improvements enhance the clinical utility of freehand 3D ultrasound by providing more reliable and precise diagnostic information.
Automatic bony structure segmentation and curvature estimation on ultrasound cervical spine images -- a feasibility study
Dec 19, 2023Abstract:The loss of cervical lordosis is a common degenerative disorder known to be associated with abnormal spinal alignment. In recent years, ultrasound (US) imaging has been widely applied in the assessment of spine deformity and has shown promising results. The objectives of this study are to automatically segment bony structures from the 3D US cervical spine image volume and to assess the cervical lordosis on the key sagittal frames. In this study, a portable ultrasound imaging system was applied to acquire cervical spine image volume. The nnU-Net was trained on to segment bony structures on the transverse images and validated by 5-fold-cross-validation. The volume data were reconstructed from the segmented image series. An energy function indicating intensity levels and integrity of bony structures was designed to extract the proxy key sagittal frames on both left and right sides for the cervical curve measurement. The mean absolute difference (MAD), standard deviation (SD) and correlation between the spine curvatures of the left and right sides were calculated for quantitative evaluation of the proposed method. The DSC value of the nnU-Net model in segmenting ROI was 0.973. For the measurement of 22 lamina curve angles, the MAD, SD and correlation between the left and right sides of the cervical spine were 3.591, 3.432 degrees and 0.926, respectively. The results indicate that our method has a high accuracy and reliability in the automatic segmentation of the cervical spine and shows the potential of diagnosing the loss of cervical lordosis using the 3D ultrasound imaging technique.
VertMatch: A Semi-supervised Framework for Vertebral Structure Detection in 3D Ultrasound Volume
Dec 28, 2022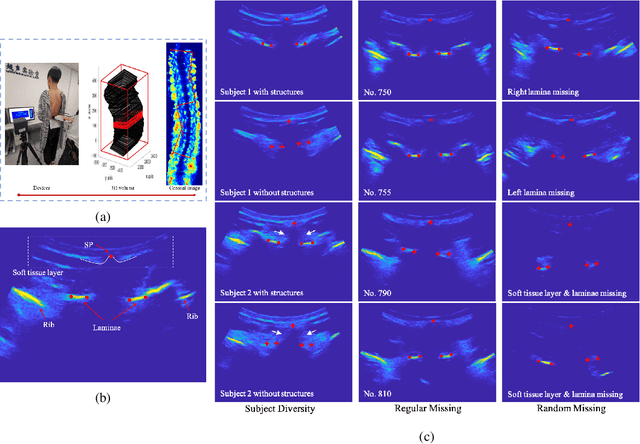

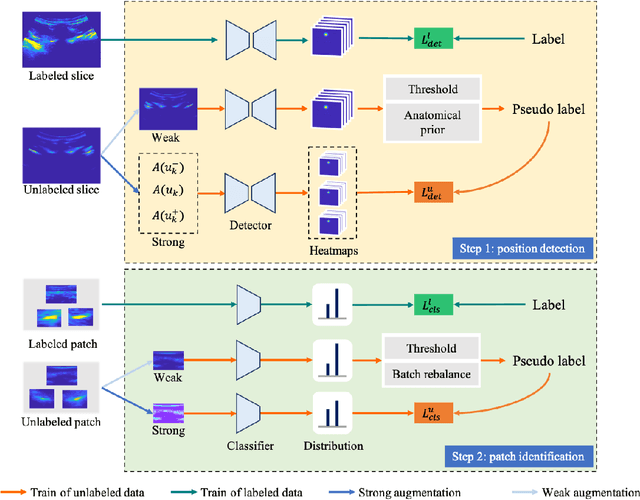
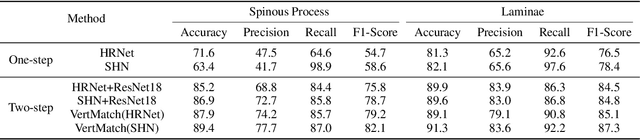
Abstract:Three-dimensional (3D) ultrasound imaging technique has been applied for scoliosis assessment, but current assessment method only uses coronal projection image and cannot illustrate the 3D deformity and vertebra rotation. The vertebra detection is essential to reveal 3D spine information, but the detection task is challenging due to complex data and limited annotations. We propose VertMatch, a two-step framework to detect vertebral structures in 3D ultrasound volume by utilizing unlabeled data in semi-supervised manner. The first step is to detect the possible positions of structures on transverse slice globally, and then the local patches are cropped based on detected positions. The second step is to distinguish whether the patches contain real vertebral structures and screen the predicted positions from the first step. VertMatch develops three novel components for semi-supervised learning: for position detection in the first step, (1) anatomical prior is used to screen pseudo labels generated from confidence threshold method; (2) multi-slice consistency is used to utilize more unlabeled data by inputting multiple adjacent slices; (3) for patch identification in the second step, the categories are rebalanced in each batch to solve imbalance problem. Experimental results demonstrate that VertMatch can detect vertebra accurately in ultrasound volume and outperforms state-of-the-art methods. VertMatch is also validated in clinical application on forty ultrasound scans, and it can be a promising approach for 3D assessment of scoliosis.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge