Siyu Lin
State Key Laboratory of Natural and Biomimetic Drugs, School of Pharmaceutical Sciences, Peking University
The Llama 4 Herd: Architecture, Training, Evaluation, and Deployment Notes
Jan 15, 2026Abstract:This document consolidates publicly reported technical details about Metas Llama 4 model family. It summarizes (i) released variants (Scout and Maverick) and the broader herd context including the previewed Behemoth teacher model, (ii) architectural characteristics beyond a high-level MoE description covering routed/shared-expert structure, early-fusion multimodality, and long-context design elements reported for Scout (iRoPE and length generalization strategies), (iii) training disclosures spanning pre-training, mid-training for long-context extension, and post-training methodology (lightweight SFT, online RL, and lightweight DPO) as described in release materials, (iv) developer-reported benchmark results for both base and instruction-tuned checkpoints, and (v) practical deployment constraints observed across major serving environments, including provider-specific context limits and quantization packaging. The manuscript also summarizes licensing obligations relevant to redistribution and derivative naming, and reviews publicly described safeguards and evaluation practices. The goal is to provide a compact technical reference for researchers and practitioners who need precise, source-backed facts about Llama 4.
Knowledge Distillation for Temporal Knowledge Graph Reasoning with Large Language Models
Jan 01, 2026Abstract:Reasoning over temporal knowledge graphs (TKGs) is fundamental to improving the efficiency and reliability of intelligent decision-making systems and has become a key technological foundation for future artificial intelligence applications. Despite recent progress, existing TKG reasoning models typically rely on large parameter sizes and intensive computation, leading to high hardware costs and energy consumption. These constraints hinder their deployment on resource-constrained, low-power, and distributed platforms that require real-time inference. Moreover, most existing model compression and distillation techniques are designed for static knowledge graphs and fail to adequately capture the temporal dependencies inherent in TKGs, often resulting in degraded reasoning performance. To address these challenges, we propose a distillation framework specifically tailored for temporal knowledge graph reasoning. Our approach leverages large language models as teacher models to guide the distillation process, enabling effective transfer of both structural and temporal reasoning capabilities to lightweight student models. By integrating large-scale public knowledge with task-specific temporal information, the proposed framework enhances the student model's ability to model temporal dynamics while maintaining a compact and efficient architecture. Extensive experiments on multiple publicly available benchmark datasets demonstrate that our method consistently outperforms strong baselines, achieving a favorable trade-off between reasoning accuracy, computational efficiency, and practical deployability.
Analytical Model of NR-V2X Mode 2 with Re-Evaluation Mechanism
Oct 31, 2025Abstract:Massive message transmissions, unpredictable aperiodic messages, and high-speed moving vehicles contribute to the complex wireless environment, resulting in inefficient resource collisions in Vehicle to Everything (V2X). In order to achieve better medium access control (MAC) layer performance, 3GPP introduced several new features in NR-V2X. One of the most important is the re-evaluation mechanism. It allows the vehicle to continuously sense resources before message transmission to avoid resource collisions. So far, only a few articles have studied the re-evaluation mechanism of NR-V2X, and they mainly focus on network simulator that do not consider variable traffic, which makes analysis and comparison difficult. In this paper, an analytical model of NR-V2X Mode 2 is established, and a message generator is constructed by using discrete time Markov chain (DTMC) to simulate the traffic pattern recommended by 3GPP advanced V2X services. Our study shows that the re-evaluation mechanism improves the reliability of NR-V2X transmission, but there are still local improvements needed to reduce latency.
Pinching-Antenna Systems (PASS) Meet Multiple Access: NOMA or OMA?
Jun 16, 2025Abstract:A fundamental two-user PASS-based communication system is considered under three MA schemes, namely non-orthogonal multiple access (NOMA), frequency division multiple access (FDMA), and time division multiple access (TDMA). For each MA scheme, a pinching beamforming optimization problem is formulated to minimize the required transmit power for satisfying users' rate requirements. For NOMA and FDMA, a two-stage algorithm is proposed, where the locations of PAs are derived sequentially by using the successive convex approximation (SCA) method and fine-turning phase adjustment. For TDMA, by leveraging the time-switching feature of PASS, the optimal pinching beamforming of each time slot is derived to maximize the served user channel gain. Numerical results are provided to show that: 1) PASS can achieve a significant performance gain over conventional antenna systems, and 2) NOMA consistently outperforms FDMA, while TDMA provides superior performance than NOMA for symmetric user rate requirements.
Computer-aided shape features extraction and regression models for predicting the ascending aortic aneurysm growth rate
Mar 04, 2025



Abstract:Objective: ascending aortic aneurysm growth prediction is still challenging in clinics. In this study, we evaluate and compare the ability of local and global shape features to predict ascending aortic aneurysm growth. Material and methods: 70 patients with aneurysm, for which two 3D acquisitions were available, are included. Following segmentation, three local shape features are computed: (1) the ratio between maximum diameter and length of the ascending aorta centerline, (2) the ratio between the length of external and internal lines on the ascending aorta and (3) the tortuosity of the ascending tract. By exploiting longitudinal data, the aneurysm growth rate is derived. Using radial basis function mesh morphing, iso-topological surface meshes are created. Statistical shape analysis is performed through unsupervised principal component analysis (PCA) and supervised partial least squares (PLS). Two types of global shape features are identified: three PCA-derived and three PLS-based shape modes. Three regression models are set for growth prediction: two based on gaussian support vector machine using local and PCA-derived global shape features; the third is a PLS linear regression model based on the related global shape features. The prediction results are assessed and the aortic shapes most prone to growth are identified. Results: the prediction root mean square error from leave-one-out cross-validation is: 0.112 mm/month, 0.083 mm/month and 0.066 mm/month for local, PCA-based and PLS-derived shape features, respectively. Aneurysms close to the root with a large initial diameter report faster growth. Conclusion: global shape features might provide an important contribution for predicting the aneurysm growth.
Efficient, gigapixel-scale, aberration-free whole slide scanner using angular ptychographic imaging with closed-form solution
Jul 29, 2024



Abstract:Whole slide imaging provides a wide field-of-view (FOV) across cross-sections of biopsy or surgery samples, significantly facilitating pathological analysis and clinical diagnosis. Such high-quality images that enable detailed visualization of cellular and tissue structures are essential for effective patient care and treatment planning. To obtain such high-quality images for pathology applications, there is a need for scanners with high spatial bandwidth products, free from aberrations, and without the requirement for z-scanning. Here we report a whole slide imaging system based on angular ptychographic imaging with a closed-form solution (WSI-APIC), which offers efficient, tens-of-gigapixels, large-FOV, aberration-free imaging. WSI-APIC utilizes oblique incoherent illumination for initial high-level segmentation, thereby bypassing unnecessary scanning of the background regions and enhancing image acquisition efficiency. A GPU-accelerated APIC algorithm analytically reconstructs phase images with effective digital aberration corrections and improved optical resolutions. Moreover, an auto-stitching technique based on scale-invariant feature transform ensures the seamless concatenation of whole slide phase images. In our experiment, WSI-APIC achieved an optical resolution of 772 nm using a 10x/0.25 NA objective lens and captures 80-gigapixel aberration-free phase images for a standard 76.2 mm x 25.4 mm microscopic slide.
Latent Chemical Space Searching for Plug-in Multi-objective Molecule Generation
Apr 10, 2024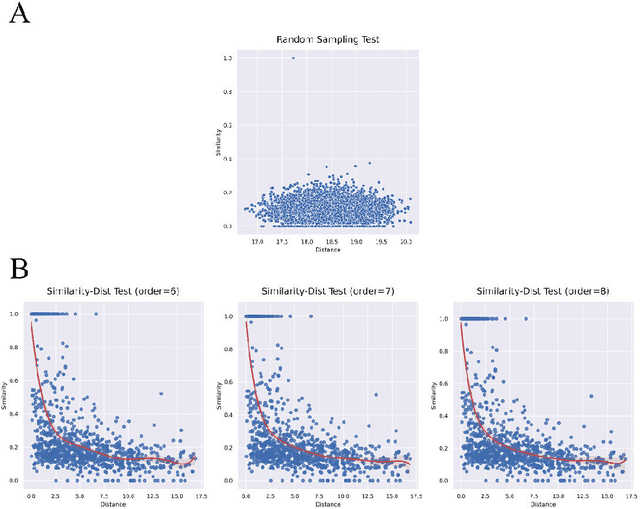
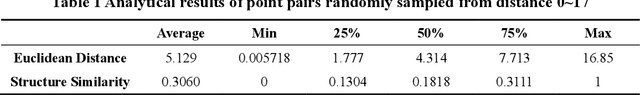
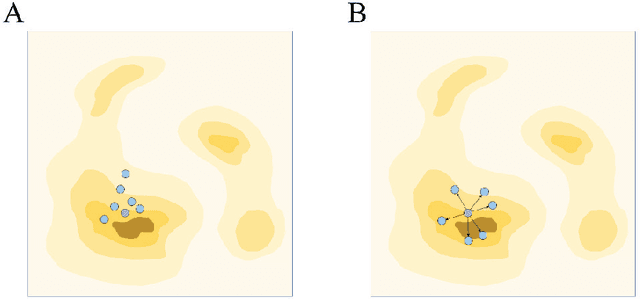
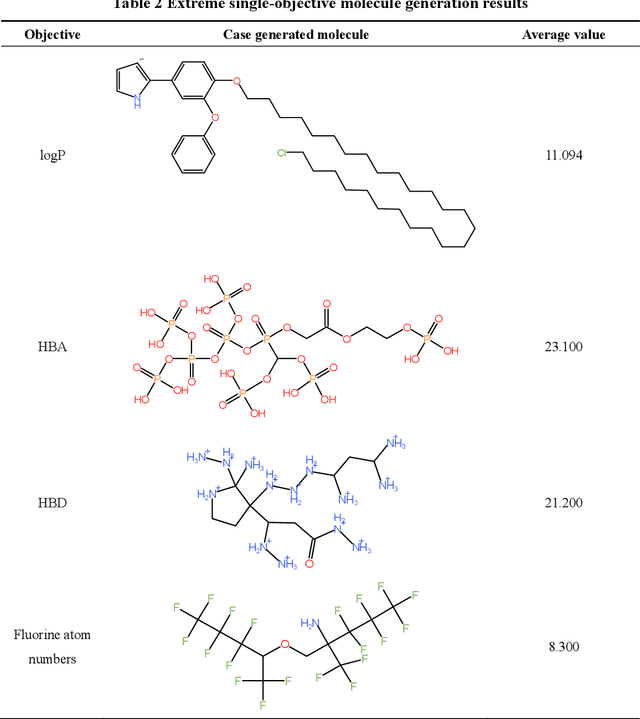
Abstract:Molecular generation, an essential method for identifying new drug structures, has been supported by advancements in machine learning and computational technology. However, challenges remain in multi-objective generation, model adaptability, and practical application in drug discovery. In this study, we developed a versatile 'plug-in' molecular generation model that incorporates multiple objectives related to target affinity, drug-likeness, and synthesizability, facilitating its application in various drug development contexts. We improved the Particle Swarm Optimization (PSO) in the context of drug discoveries, and identified PSO-ENP as the optimal variant for multi-objective molecular generation and optimization through comparative experiments. The model also incorporates a novel target-ligand affinity predictor, enhancing the model's utility by supporting three-dimensional information and improving synthetic feasibility. Case studies focused on generating and optimizing drug-like big marine natural products were performed, underscoring PSO-ENP's effectiveness and demonstrating its considerable potential for practical drug discovery applications.
FPM-INR: Fourier ptychographic microscopy image stack reconstruction using implicit neural representations
Oct 31, 2023
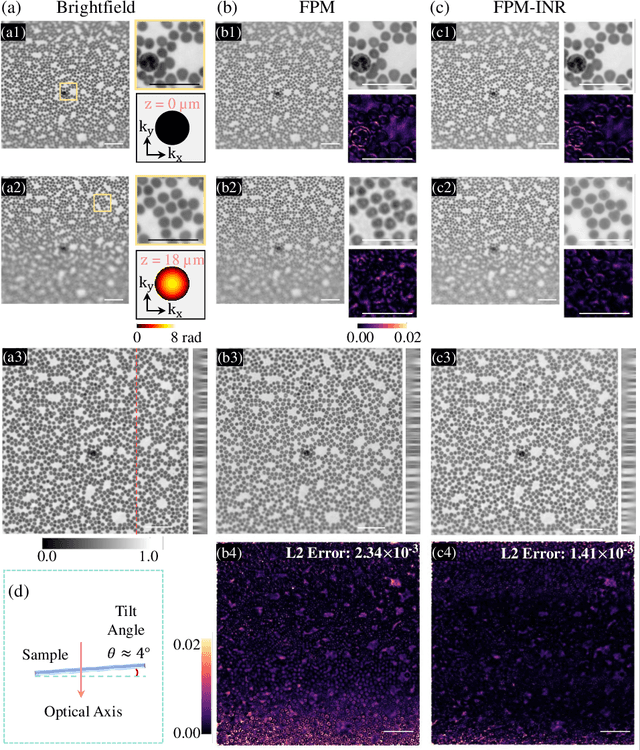
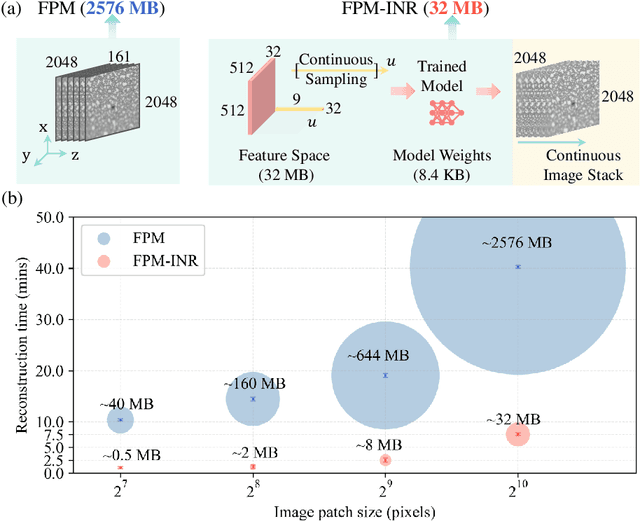
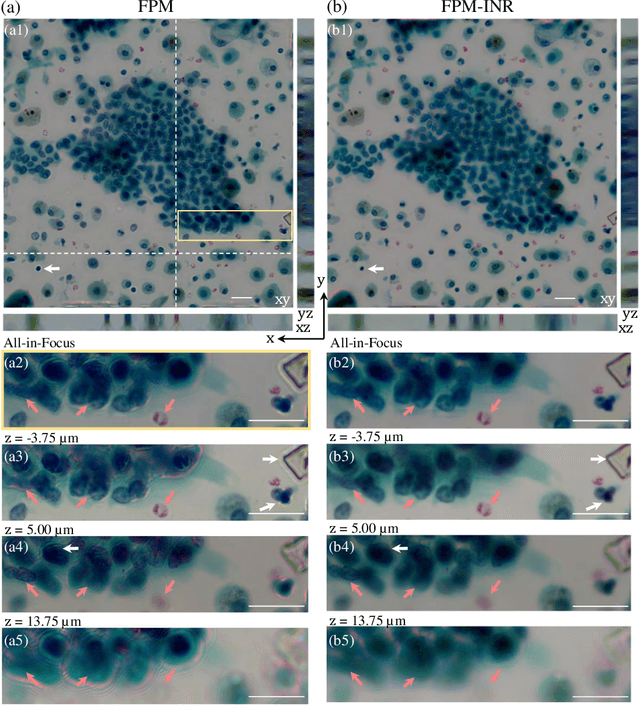
Abstract:Image stacks provide invaluable 3D information in various biological and pathological imaging applications. Fourier ptychographic microscopy (FPM) enables reconstructing high-resolution, wide field-of-view image stacks without z-stack scanning, thus significantly accelerating image acquisition. However, existing FPM methods take tens of minutes to reconstruct and gigabytes of memory to store a high-resolution volumetric scene, impeding fast gigapixel-scale remote digital pathology. While deep learning approaches have been explored to address this challenge, existing methods poorly generalize to novel datasets and can produce unreliable hallucinations. This work presents FPM-INR, a compact and efficient framework that integrates physics-based optical models with implicit neural representations (INR) to represent and reconstruct FPM image stacks. FPM-INR is agnostic to system design or sample types and does not require external training data. In our demonstrated experiments, FPM-INR substantially outperforms traditional FPM algorithms with up to a 25-fold increase in speed and an 80-fold reduction in memory usage for continuous image stack representations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge