Haowen Zhou
Siyu
Digital Modeling of Spatial Pathway Activity from Histology Reveals Tumor Microenvironment Heterogeneity
Dec 18, 2025
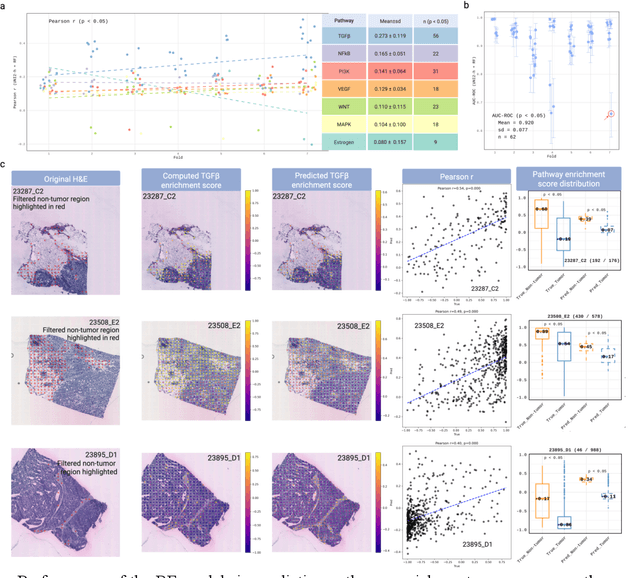
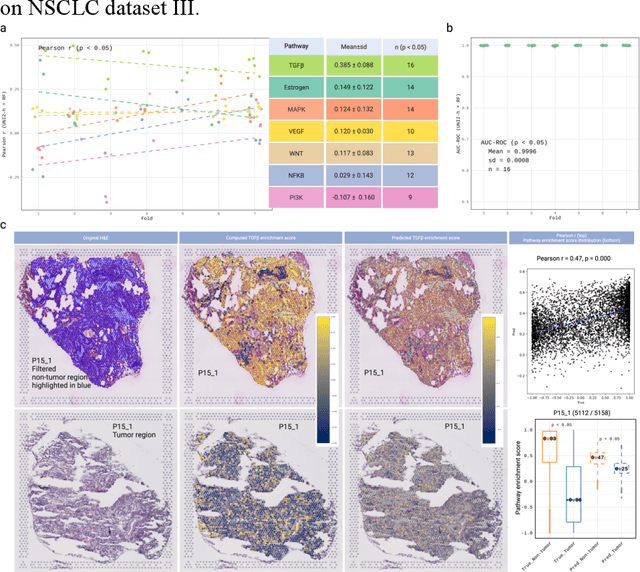
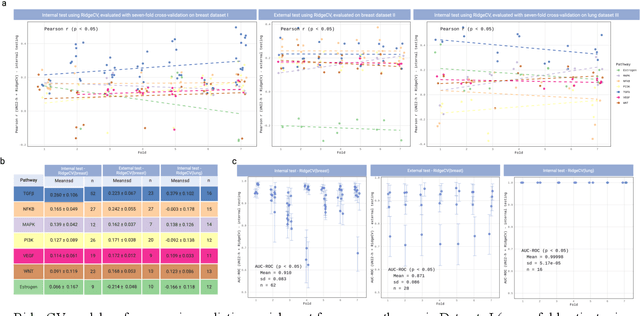
Abstract:Spatial transcriptomics (ST) enables simultaneous mapping of tissue morphology and spatially resolved gene expression, offering unique opportunities to study tumor microenvironment heterogeneity. Here, we introduce a computational framework that predicts spatial pathway activity directly from hematoxylin-and-eosin-stained histology images at microscale resolution 55 and 100 um. Using image features derived from a computational pathology foundation model, we found that TGFb signaling was the most accurately predicted pathway across three independent breast and lung cancer ST datasets. In 87-88% of reliably predicted cases, the resulting spatial TGFb activity maps reflected the expected contrast between tumor and adjacent non-tumor regions, consistent with the known role of TGFb in regulating interactions within the tumor microenvironment. Notably, linear and nonlinear predictive models performed similarly, suggesting that image features may relate to pathway activity in a predominantly linear fashion or that nonlinear structure is small relative to measurement noise. These findings demonstrate that features extracted from routine histopathology may recover spatially coherent and biologically interpretable pathway patterns, offering a scalable strategy for integrating image-based inference with ST information in tumor microenvironment studies.
Impact of Stain Variation and Color Normalization for Prognostic Predictions in Pathology
Sep 12, 2024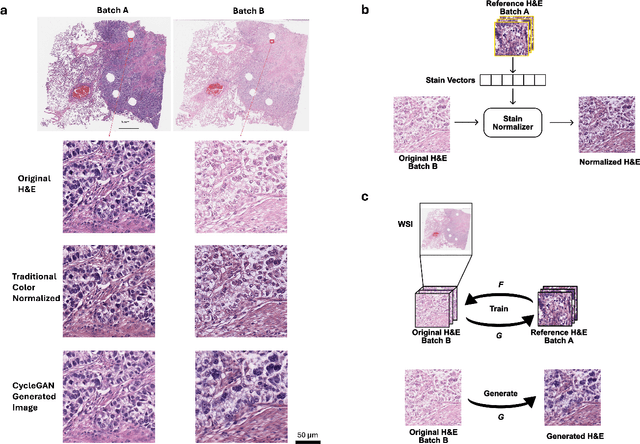
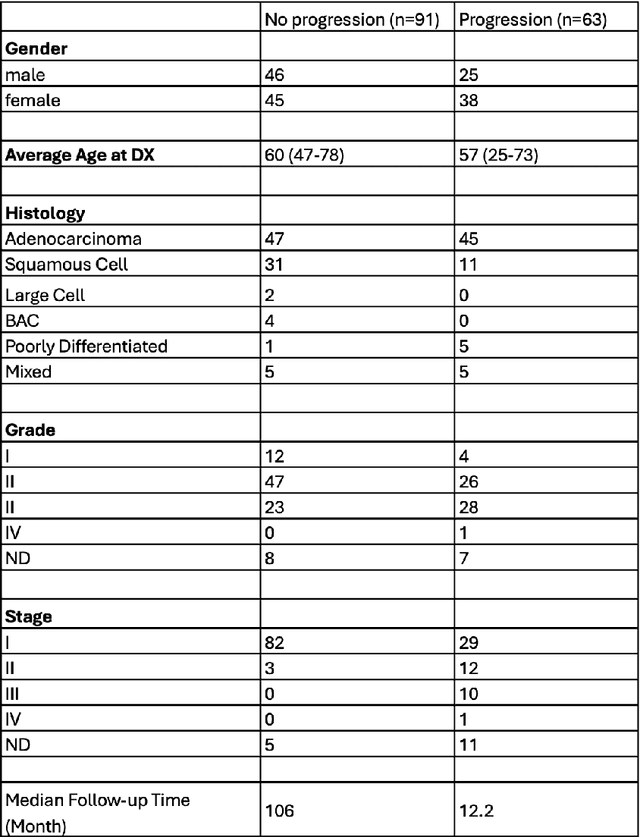
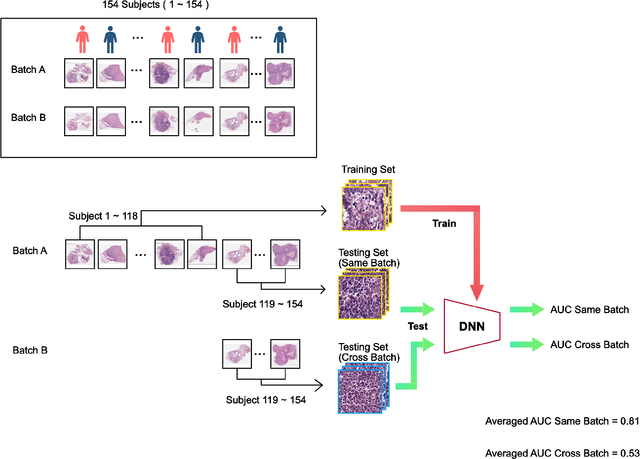
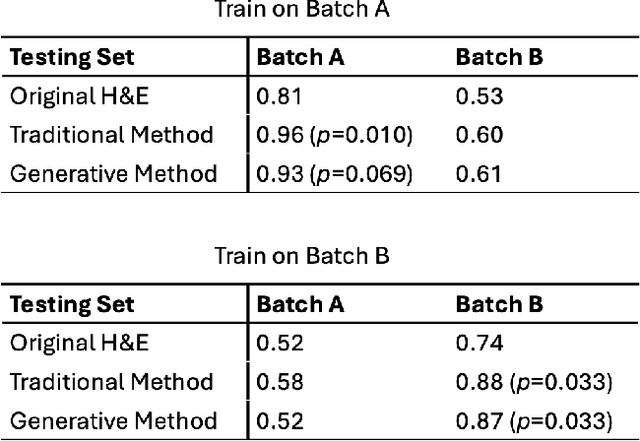
Abstract:In recent years, deep neural networks (DNNs) have demonstrated remarkable performance in pathology applications, potentially even outperforming expert pathologists due to their ability to learn subtle features from large datasets. One complication in preparing digital pathology datasets for DNN tasks is variation in tinctorial qualities. A common way to address this is to perform stain normalization on the images. In this study, we show that a well-trained DNN model trained on one batch of histological slides failed to generalize to another batch prepared at a different time from the same tissue blocks, even when stain normalization methods were applied. This study used sample data from a previously reported DNN that was able to identify patients with early stage non-small cell lung cancer (NSCLC) whose tumors did and did not metastasize, with high accuracy, based on training and then testing of digital images from H&E stained primary tumor tissue sections processed at the same time. In this study we obtained a new series of histologic slides from the adjacent recuts of same tissue blocks processed in the same lab but at a different time. We found that the DNN trained on the either batch of slides/images was unable to generalize and failed to predict progression in the other batch of slides/images (AUC_cross-batch = 0.52 - 0.53 compared to AUC_same-batch = 0.74 - 0.81). The failure to generalize did not improve even when the tinctorial difference correction were made through either traditional color-tuning or stain normalization with the help of a Cycle Generative Adversarial Network (CycleGAN) process. This highlights the need to develop an entirely new way to process and collect consistent microscopy images from histologic slides that can be used to both train and allow for the general application of predictive DNN algorithms.
Efficient, gigapixel-scale, aberration-free whole slide scanner using angular ptychographic imaging with closed-form solution
Jul 29, 2024



Abstract:Whole slide imaging provides a wide field-of-view (FOV) across cross-sections of biopsy or surgery samples, significantly facilitating pathological analysis and clinical diagnosis. Such high-quality images that enable detailed visualization of cellular and tissue structures are essential for effective patient care and treatment planning. To obtain such high-quality images for pathology applications, there is a need for scanners with high spatial bandwidth products, free from aberrations, and without the requirement for z-scanning. Here we report a whole slide imaging system based on angular ptychographic imaging with a closed-form solution (WSI-APIC), which offers efficient, tens-of-gigapixels, large-FOV, aberration-free imaging. WSI-APIC utilizes oblique incoherent illumination for initial high-level segmentation, thereby bypassing unnecessary scanning of the background regions and enhancing image acquisition efficiency. A GPU-accelerated APIC algorithm analytically reconstructs phase images with effective digital aberration corrections and improved optical resolutions. Moreover, an auto-stitching technique based on scale-invariant feature transform ensures the seamless concatenation of whole slide phase images. In our experiment, WSI-APIC achieved an optical resolution of 772 nm using a 10x/0.25 NA objective lens and captures 80-gigapixel aberration-free phase images for a standard 76.2 mm x 25.4 mm microscopic slide.
Length-scale study in deep learning prediction for non-small cell lung cancer brain metastasis
Jun 01, 2024Abstract:Deep learning assisted digital pathology has the potential to impact clinical practice in significant ways. In recent studies, deep neural network (DNN) enabled analysis outperforms human pathologists. Increasing sizes and complexity of the DNN architecture generally improves performance at the cost of DNN's explainability. For pathology, this lack of DNN explainability is particularly problematic as it hinders the broader clinical interpretation of the pathology features that may provide physiological disease insights. To better assess the features that DNN uses in developing predictive algorithms to interpret digital microscopic images, we sought to understand the role of resolution and tissue scale and here describe a novel method for studying the predictive feature length-scale that underpins a DNN's predictive power. We applied the method to study a DNN's predictive capability in the case example of brain metastasis prediction from early-stage non-small-cell lung cancer biopsy slides. The study highlights the DNN attention in the brain metastasis prediction targeting both cellular scale (resolution) and tissue scale features on H&E-stained histological whole slide images. At the cellular scale, we see that DNN's predictive power is progressively increased at higher resolution (i.e., lower resolvable feature length) and is largely lost when the resolvable feature length is longer than 5 microns. In addition, DNN uses more macro-scale features (maximal feature length) associated with tissue organization/architecture and is optimized when assessing visual fields larger than 41 microns. This study for the first time demonstrates the length-scale requirements necessary for optimal DNN learning on digital whole slide images.
Single-shot volumetric fluorescence imaging with neural fields
May 16, 2024Abstract:Single-shot volumetric fluorescence (SVF) imaging offers a significant advantage over traditional imaging methods that require scanning across multiple axial planes as it can capture biological processes with high temporal resolution across a large field of view. Existing SVF imaging methods often require large, complex point spread functions (PSFs) to meet the multiplexing requirements of compressed sensing, which limits the signal-to-noise ratio, resolution and/or field of view. In this paper, we introduce the QuadraPol PSF combined with neural fields, a novel approach for SVF imaging. This method utilizes a cost-effective custom polarizer at the back focal plane and a polarization camera to detect fluorescence, effectively encoding the 3D scene within a compact PSF without depth ambiguity. Additionally, we propose a reconstruction algorithm based on the neural fields technique that addresses the inaccuracies of phase retrieval methods used to correct imaging system aberrations. This algorithm combines the accuracy of experimental PSFs with the long depth of field of computationally generated retrieved PSFs. QuadraPol PSF, combined with neural fields, significantly reduces the acquisition time of a conventional fluorescence microscope by approximately 20 times and captures a 100 mm$^3$ cubic volume in one shot. We validate the effectiveness of both our hardware and algorithm through all-in-focus imaging of bacterial colonies on sand surfaces and visualization of plant root morphology. Our approach offers a powerful tool for advancing biological research and ecological studies.
FPM-INR: Fourier ptychographic microscopy image stack reconstruction using implicit neural representations
Oct 31, 2023
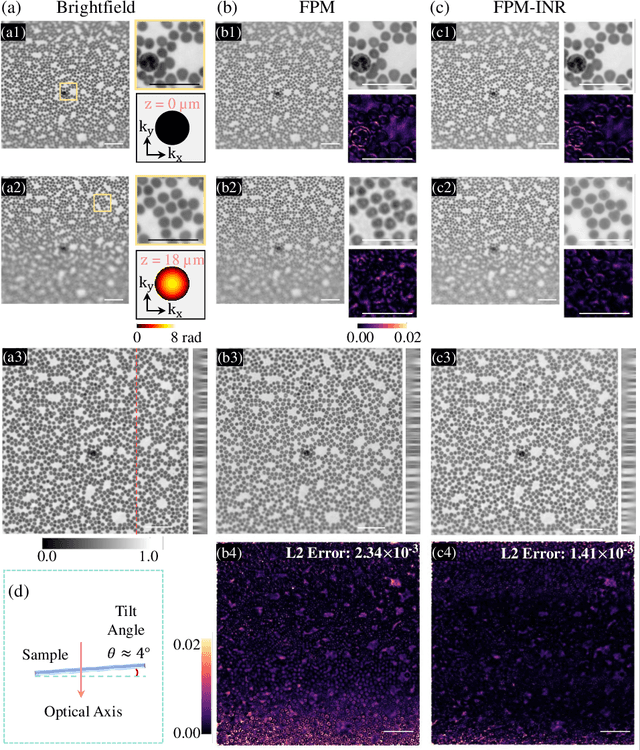
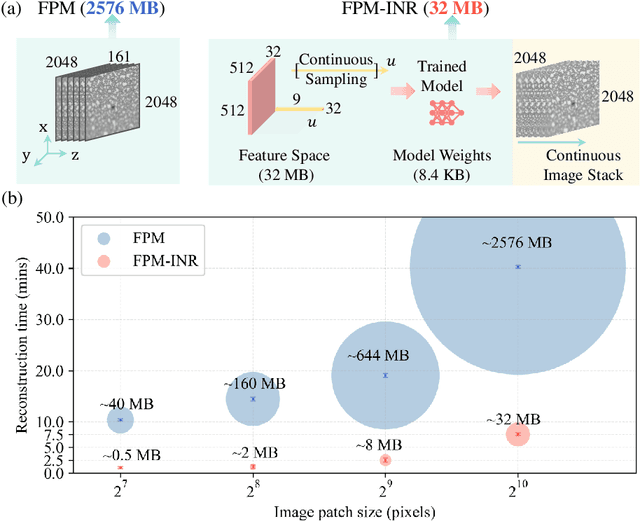
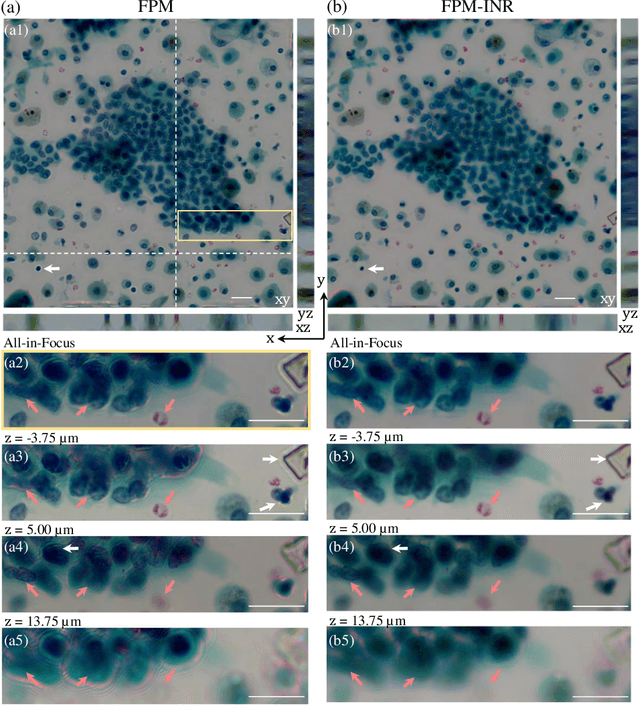
Abstract:Image stacks provide invaluable 3D information in various biological and pathological imaging applications. Fourier ptychographic microscopy (FPM) enables reconstructing high-resolution, wide field-of-view image stacks without z-stack scanning, thus significantly accelerating image acquisition. However, existing FPM methods take tens of minutes to reconstruct and gigabytes of memory to store a high-resolution volumetric scene, impeding fast gigapixel-scale remote digital pathology. While deep learning approaches have been explored to address this challenge, existing methods poorly generalize to novel datasets and can produce unreliable hallucinations. This work presents FPM-INR, a compact and efficient framework that integrates physics-based optical models with implicit neural representations (INR) to represent and reconstruct FPM image stacks. FPM-INR is agnostic to system design or sample types and does not require external training data. In our demonstrated experiments, FPM-INR substantially outperforms traditional FPM algorithms with up to a 25-fold increase in speed and an 80-fold reduction in memory usage for continuous image stack representations.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge