Mark Watson
Digital Modeling of Spatial Pathway Activity from Histology Reveals Tumor Microenvironment Heterogeneity
Dec 18, 2025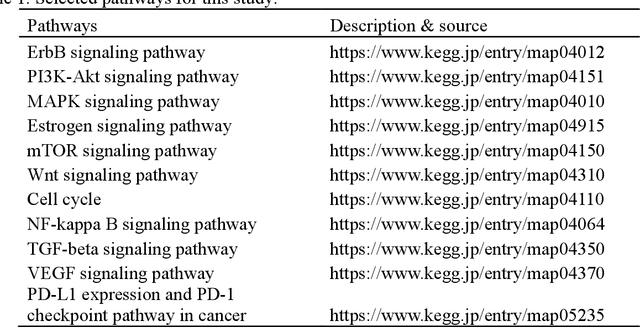
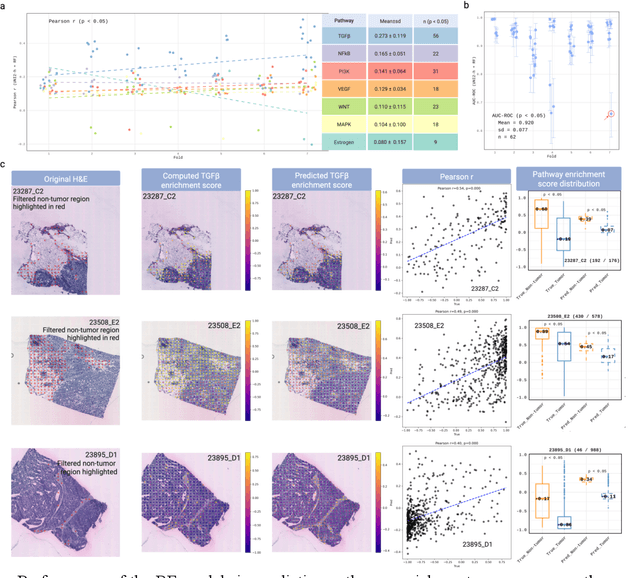
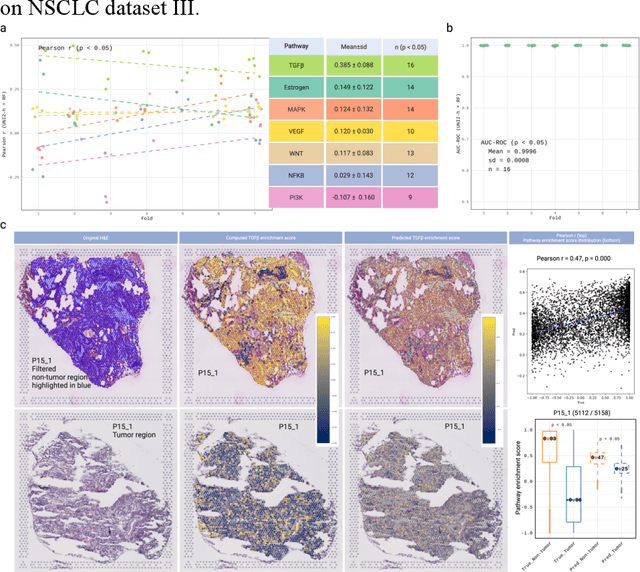
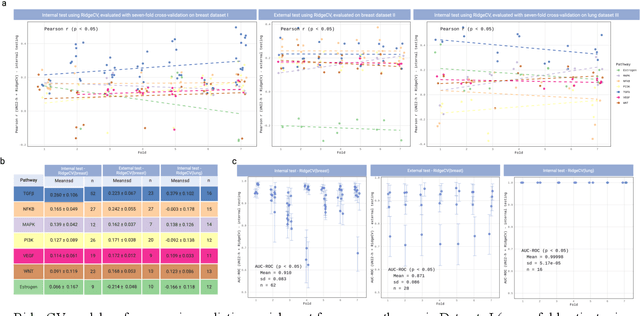
Abstract:Spatial transcriptomics (ST) enables simultaneous mapping of tissue morphology and spatially resolved gene expression, offering unique opportunities to study tumor microenvironment heterogeneity. Here, we introduce a computational framework that predicts spatial pathway activity directly from hematoxylin-and-eosin-stained histology images at microscale resolution 55 and 100 um. Using image features derived from a computational pathology foundation model, we found that TGFb signaling was the most accurately predicted pathway across three independent breast and lung cancer ST datasets. In 87-88% of reliably predicted cases, the resulting spatial TGFb activity maps reflected the expected contrast between tumor and adjacent non-tumor regions, consistent with the known role of TGFb in regulating interactions within the tumor microenvironment. Notably, linear and nonlinear predictive models performed similarly, suggesting that image features may relate to pathway activity in a predominantly linear fashion or that nonlinear structure is small relative to measurement noise. These findings demonstrate that features extracted from routine histopathology may recover spatially coherent and biologically interpretable pathway patterns, offering a scalable strategy for integrating image-based inference with ST information in tumor microenvironment studies.
Impact of Stain Variation and Color Normalization for Prognostic Predictions in Pathology
Sep 12, 2024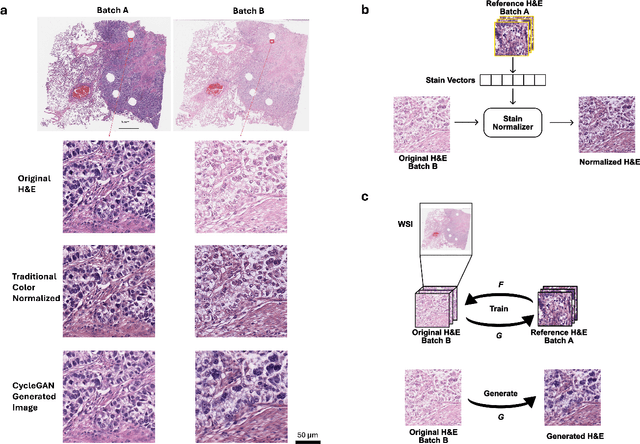
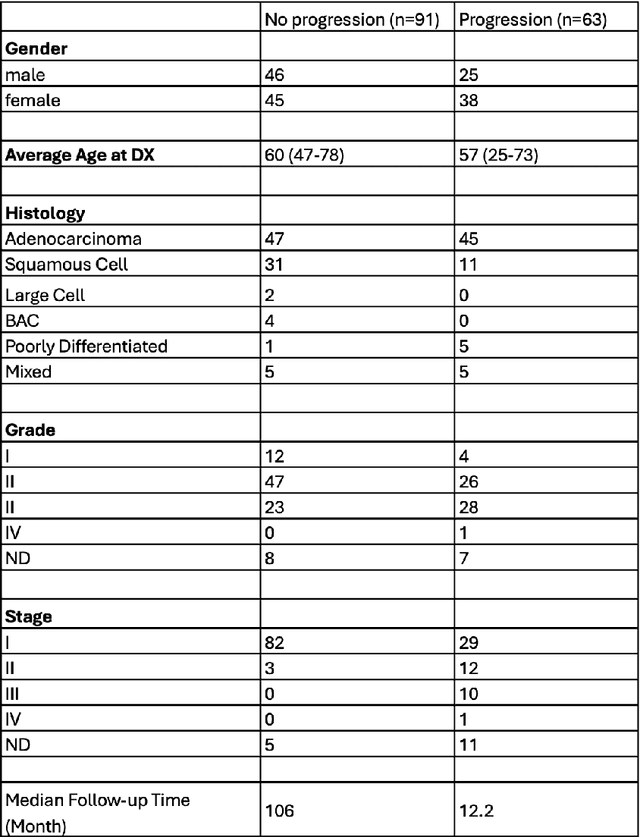
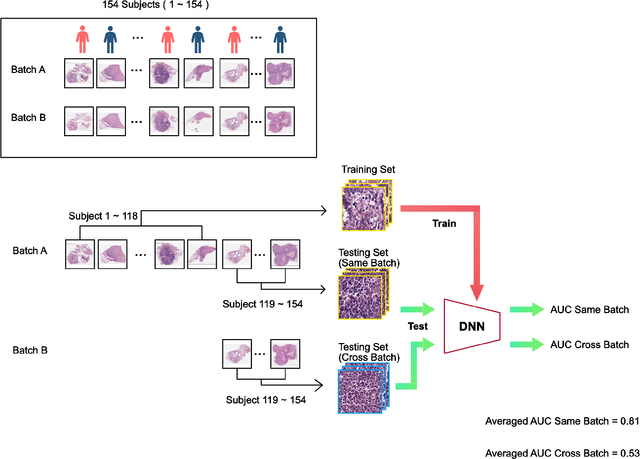
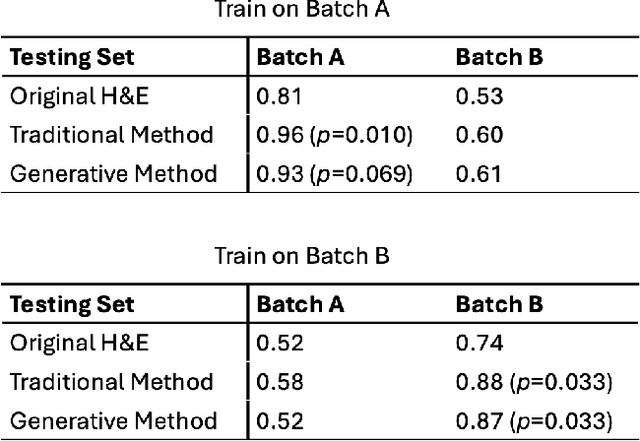
Abstract:In recent years, deep neural networks (DNNs) have demonstrated remarkable performance in pathology applications, potentially even outperforming expert pathologists due to their ability to learn subtle features from large datasets. One complication in preparing digital pathology datasets for DNN tasks is variation in tinctorial qualities. A common way to address this is to perform stain normalization on the images. In this study, we show that a well-trained DNN model trained on one batch of histological slides failed to generalize to another batch prepared at a different time from the same tissue blocks, even when stain normalization methods were applied. This study used sample data from a previously reported DNN that was able to identify patients with early stage non-small cell lung cancer (NSCLC) whose tumors did and did not metastasize, with high accuracy, based on training and then testing of digital images from H&E stained primary tumor tissue sections processed at the same time. In this study we obtained a new series of histologic slides from the adjacent recuts of same tissue blocks processed in the same lab but at a different time. We found that the DNN trained on the either batch of slides/images was unable to generalize and failed to predict progression in the other batch of slides/images (AUC_cross-batch = 0.52 - 0.53 compared to AUC_same-batch = 0.74 - 0.81). The failure to generalize did not improve even when the tinctorial difference correction were made through either traditional color-tuning or stain normalization with the help of a Cycle Generative Adversarial Network (CycleGAN) process. This highlights the need to develop an entirely new way to process and collect consistent microscopy images from histologic slides that can be used to both train and allow for the general application of predictive DNN algorithms.
Length-scale study in deep learning prediction for non-small cell lung cancer brain metastasis
Jun 01, 2024Abstract:Deep learning assisted digital pathology has the potential to impact clinical practice in significant ways. In recent studies, deep neural network (DNN) enabled analysis outperforms human pathologists. Increasing sizes and complexity of the DNN architecture generally improves performance at the cost of DNN's explainability. For pathology, this lack of DNN explainability is particularly problematic as it hinders the broader clinical interpretation of the pathology features that may provide physiological disease insights. To better assess the features that DNN uses in developing predictive algorithms to interpret digital microscopic images, we sought to understand the role of resolution and tissue scale and here describe a novel method for studying the predictive feature length-scale that underpins a DNN's predictive power. We applied the method to study a DNN's predictive capability in the case example of brain metastasis prediction from early-stage non-small-cell lung cancer biopsy slides. The study highlights the DNN attention in the brain metastasis prediction targeting both cellular scale (resolution) and tissue scale features on H&E-stained histological whole slide images. At the cellular scale, we see that DNN's predictive power is progressively increased at higher resolution (i.e., lower resolvable feature length) and is largely lost when the resolvable feature length is longer than 5 microns. In addition, DNN uses more macro-scale features (maximal feature length) associated with tissue organization/architecture and is optimized when assessing visual fields larger than 41 microns. This study for the first time demonstrates the length-scale requirements necessary for optimal DNN learning on digital whole slide images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge