Simon Bing
Sanity Checking Causal Representation Learning on a Simple Real-World System
Feb 27, 2025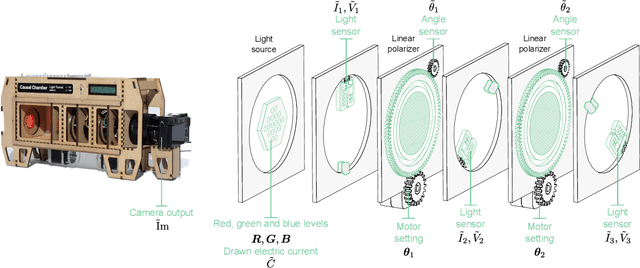
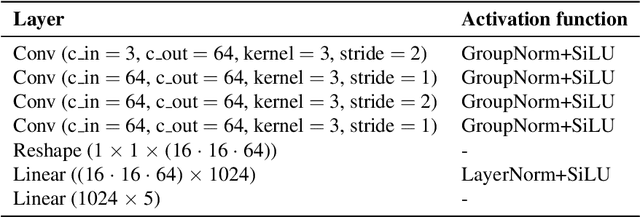
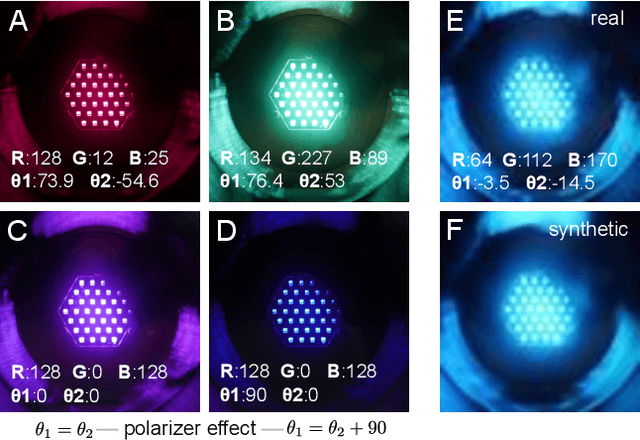
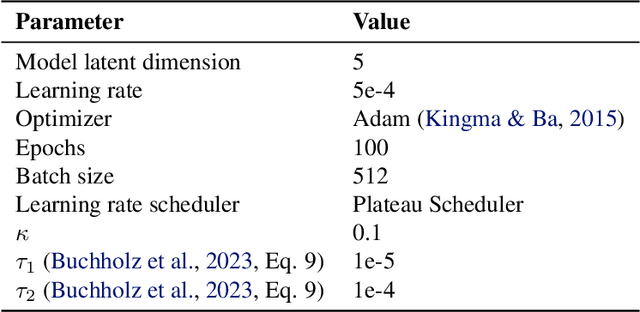
Abstract:We evaluate methods for causal representation learning (CRL) on a simple, real-world system where these methods are expected to work. The system consists of a controlled optical experiment specifically built for this purpose, which satisfies the core assumptions of CRL and where the underlying causal factors (the inputs to the experiment) are known, providing a ground truth. We select methods representative of different approaches to CRL and find that they all fail to recover the underlying causal factors. To understand the failure modes of the evaluated algorithms, we perform an ablation on the data by substituting the real data-generating process with a simpler synthetic equivalent. The results reveal a reproducibility problem, as most methods already fail on this synthetic ablation despite its simple data-generating process. Additionally, we observe that common assumptions on the mixing function are crucial for the performance of some of the methods but do not hold in the real data. Our efforts highlight the contrast between the theoretical promise of the state of the art and the challenges in its application. We hope the benchmark serves as a simple, real-world sanity check to further develop and validate methodology, bridging the gap towards CRL methods that work in practice. We make all code and datasets publicly available at github.com/simonbing/CRLSanityCheck
Invariance & Causal Representation Learning: Prospects and Limitations
Dec 06, 2023Abstract:In causal models, a given mechanism is assumed to be invariant to changes of other mechanisms. While this principle has been utilized for inference in settings where the causal variables are observed, theoretical insights when the variables of interest are latent are largely missing. We assay the connection between invariance and causal representation learning by establishing impossibility results which show that invariance alone is insufficient to identify latent causal variables. Together with practical considerations, we use these theoretical findings to highlight the need for additional constraints in order to identify representations by exploiting invariance.
Identifying Linearly-Mixed Causal Representations from Multi-Node Interventions
Nov 05, 2023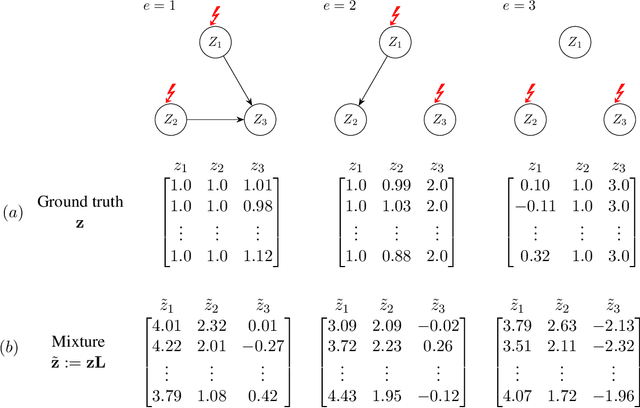


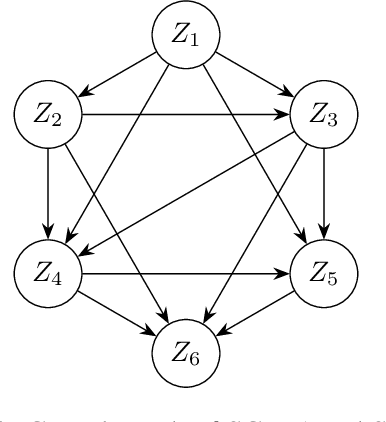
Abstract:The task of inferring high-level causal variables from low-level observations, commonly referred to as causal representation learning, is fundamentally underconstrained. As such, recent works to address this problem focus on various assumptions that lead to identifiability of the underlying latent causal variables. A large corpus of these preceding approaches consider multi-environment data collected under different interventions on the causal model. What is common to virtually all of these works is the restrictive assumption that in each environment, only a single variable is intervened on. In this work, we relax this assumption and provide the first identifiability result for causal representation learning that allows for multiple variables to be targeted by an intervention within one environment. Our approach hinges on a general assumption on the coverage and diversity of interventions across environments, which also includes the shared assumption of single-node interventions of previous works. The main idea behind our approach is to exploit the trace that interventions leave on the variance of the ground truth causal variables and regularizing for a specific notion of sparsity with respect to this trace. In addition to and inspired by our theoretical contributions, we present a practical algorithm to learn causal representations from multi-node interventional data and provide empirical evidence that validates our identifiability results.
Conditional Generation of Medical Time Series for Extrapolation to Underrepresented Populations
Jan 20, 2022



Abstract:The widespread adoption of electronic health records (EHRs) and subsequent increased availability of longitudinal healthcare data has led to significant advances in our understanding of health and disease with direct and immediate impact on the development of new diagnostics and therapeutic treatment options. However, access to EHRs is often restricted due to their perceived sensitive nature and associated legal concerns, and the cohorts therein typically are those seen at a specific hospital or network of hospitals and therefore not representative of the wider population of patients. Here, we present HealthGen, a new approach for the conditional generation of synthetic EHRs that maintains an accurate representation of real patient characteristics, temporal information and missingness patterns. We demonstrate experimentally that HealthGen generates synthetic cohorts that are significantly more faithful to real patient EHRs than the current state-of-the-art, and that augmenting real data sets with conditionally generated cohorts of underrepresented subpopulations of patients can significantly enhance the generalisability of models derived from these data sets to different patient populations. Synthetic conditionally generated EHRs could help increase the accessibility of longitudinal healthcare data sets and improve the generalisability of inferences made from these data sets to underrepresented populations.
On Disentanglement in Gaussian Process Variational Autoencoders
Feb 10, 2021



Abstract:Complex multivariate time series arise in many fields, ranging from computer vision to robotics or medicine. Often we are interested in the independent underlying factors that give rise to the high-dimensional data we are observing. While many models have been introduced to learn such disentangled representations, only few attempt to explicitly exploit the structure of sequential data. We investigate the disentanglement properties of Gaussian process variational autoencoders, a class of models recently introduced that have been successful in different tasks on time series data. Our model exploits the temporal structure of the data by modeling each latent channel with a GP prior and employing a structured variational distribution that can capture dependencies in time. We demonstrate the competitiveness of our approach against state-of-the-art unsupervised and weakly-supervised disentanglement methods on a benchmark task. Moreover, we provide evidence that we can learn meaningful disentangled representations on real-world medical time series data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge