Sebastian Joseph
Decide less, communicate more: On the construct validity of end-to-end fact-checking in medicine
Jun 25, 2025Abstract:Technological progress has led to concrete advancements in tasks that were regarded as challenging, such as automatic fact-checking. Interest in adopting these systems for public health and medicine has grown due to the high-stakes nature of medical decisions and challenges in critically appraising a vast and diverse medical literature. Evidence-based medicine connects to every individual, and yet the nature of it is highly technical, rendering the medical literacy of majority users inadequate to sufficiently navigate the domain. Such problems with medical communication ripens the ground for end-to-end fact-checking agents: check a claim against current medical literature and return with an evidence-backed verdict. And yet, such systems remain largely unused. To understand this, we present the first study examining how clinical experts verify real claims from social media by synthesizing medical evidence. In searching for this upper-bound, we reveal fundamental challenges in end-to-end fact-checking when applied to medicine: Difficulties connecting claims in the wild to scientific evidence in the form of clinical trials; ambiguities in underspecified claims mixed with mismatched intentions; and inherently subjective veracity labels. We argue that fact-checking should be approached and evaluated as an interactive communication problem, rather than an end-to-end process.
InfoLossQA: Characterizing and Recovering Information Loss in Text Simplification
Jan 29, 2024



Abstract:Text simplification aims to make technical texts more accessible to laypeople but often results in deletion of information and vagueness. This work proposes InfoLossQA, a framework to characterize and recover simplification-induced information loss in form of question-and-answer (QA) pairs. Building on the theory of Question Under Discussion, the QA pairs are designed to help readers deepen their knowledge of a text. We conduct a range of experiments with this framework. First, we collect a dataset of 1,000 linguist-curated QA pairs derived from 104 LLM simplifications of scientific abstracts of medical studies. Our analyses of this data reveal that information loss occurs frequently, and that the QA pairs give a high-level overview of what information was lost. Second, we devise two methods for this task: end-to-end prompting of open-source and commercial language models, and a natural language inference pipeline. With a novel evaluation framework considering the correctness of QA pairs and their linguistic suitability, our expert evaluation reveals that models struggle to reliably identify information loss and applying similar standards as humans at what constitutes information loss.
Multilingual Simplification of Medical Texts
May 23, 2023



Abstract:Automated text simplification aims to produce simple versions of complex texts. This task is especially useful in the medical domain, where the latest medical findings are typically communicated via complex and technical articles. This creates barriers for laypeople seeking access to up-to-date medical findings, consequently impeding progress on health literacy. Most existing work on medical text simplification has focused on monolingual settings, with the result that such evidence would be available only in just one language (most often, English). This work addresses this limitation via multilingual simplification, i.e., directly simplifying complex texts into simplified texts in multiple languages. We introduce MultiCochrane, the first sentence-aligned multilingual text simplification dataset for the medical domain in four languages: English, Spanish, French, and Farsi. We evaluate fine-tuned and zero-shot models across these languages, with extensive human assessments and analyses. Although models can now generate viable simplified texts, we identify outstanding challenges that this dataset might be used to address.
Summarizing, Simplifying, and Synthesizing Medical Evidence Using GPT-3 (with Varying Success)
May 11, 2023

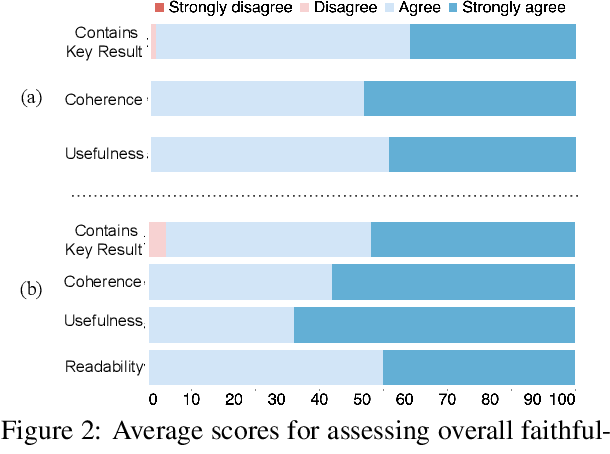

Abstract:Large language models, particularly GPT-3, are able to produce high quality summaries of general domain news articles in few- and zero-shot settings. However, it is unclear if such models are similarly capable in more specialized, high-stakes domains such as biomedicine. In this paper, we enlist domain experts (individuals with medical training) to evaluate summaries of biomedical articles generated by GPT-3, given zero supervision. We consider both single- and multi-document settings. In the former, GPT-3 is tasked with generating regular and plain-language summaries of articles describing randomized controlled trials; in the latter, we assess the degree to which GPT-3 is able to \emph{synthesize} evidence reported across a collection of articles. We design an annotation scheme for evaluating model outputs, with an emphasis on assessing the factual accuracy of generated summaries. We find that while GPT-3 is able to summarize and simplify single biomedical articles faithfully, it struggles to provide accurate aggregations of findings over multiple documents. We release all data and annotations used in this work.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge