Sarina Thomas
EchoNarrator: Generating natural text explanations for ejection fraction predictions
Oct 31, 2024Abstract:Ejection fraction (EF) of the left ventricle (LV) is considered as one of the most important measurements for diagnosing acute heart failure and can be estimated during cardiac ultrasound acquisition. While recent successes in deep learning research successfully estimate EF values, the proposed models often lack an explanation for the prediction. However, providing clear and intuitive explanations for clinical measurement predictions would increase the trust of cardiologists in these models. In this paper, we explore predicting EF measurements with Natural Language Explanation (NLE). We propose a model that in a single forward pass combines estimation of the LV contour over multiple frames, together with a set of modules and routines for computing various motion and shape attributes that are associated with ejection fraction. It then feeds the attributes into a large language model to generate text that helps to explain the network's outcome in a human-like manner. We provide experimental evaluation of our explanatory output, as well as EF prediction, and show that our model can provide EF comparable to state-of-the-art together with meaningful and accurate natural language explanation to the prediction. The project page can be found at https://github.com/guybenyosef/EchoNarrator .
Graph Convolutional Neural Networks for Automated Echocardiography View Recognition: A Holistic Approach
Mar 01, 2024Abstract:To facilitate diagnosis on cardiac ultrasound (US), clinical practice has established several standard views of the heart, which serve as reference points for diagnostic measurements and define viewports from which images are acquired. Automatic view recognition involves grouping those images into classes of standard views. Although deep learning techniques have been successful in achieving this, they still struggle with fully verifying the suitability of an image for specific measurements due to factors like the correct location, pose, and potential occlusions of cardiac structures. Our approach goes beyond view classification and incorporates a 3D mesh reconstruction of the heart that enables several more downstream tasks, like segmentation and pose estimation. In this work, we explore learning 3D heart meshes via graph convolutions, using similar techniques to learn 3D meshes in natural images, such as human pose estimation. As the availability of fully annotated 3D images is limited, we generate synthetic US images from 3D meshes by training an adversarial denoising diffusion model. Experiments were conducted on synthetic and clinical cases for view recognition and structure detection. The approach yielded good performance on synthetic images and, despite being exclusively trained on synthetic data, it already showed potential when applied to clinical images. With this proof-of-concept, we aim to demonstrate the benefits of graphs to improve cardiac view recognition that can ultimately lead to better efficiency in cardiac diagnosis.
Towards Robust Cardiac Segmentation using Graph Convolutional Networks
Oct 02, 2023



Abstract:Fully automatic cardiac segmentation can be a fast and reproducible method to extract clinical measurements from an echocardiography examination. The U-Net architecture is the current state-of-the-art deep learning architecture for medical segmentation and can segment cardiac structures in real-time with average errors comparable to inter-observer variability. However, this architecture still generates large outliers that are often anatomically incorrect. This work uses the concept of graph convolutional neural networks that predict the contour points of the structures of interest instead of labeling each pixel. We propose a graph architecture that uses two convolutional rings based on cardiac anatomy and show that this eliminates anatomical incorrect multi-structure segmentations on the publicly available CAMUS dataset. Additionally, this work contributes with an ablation study on the graph convolutional architecture and an evaluation of clinical measurements on the clinical HUNT4 dataset. Finally, we propose to use the inter-model agreement of the U-Net and the graph network as a predictor of both the input and segmentation quality. We show this predictor can detect out-of-distribution and unsuitable input images in real-time. Source code is available online: https://github.com/gillesvntnu/GCN_multistructure
Shape-based pose estimation for automatic standard views of the knee
May 26, 2023



Abstract:Surgical treatment of complicated knee fractures is guided by real-time imaging using a mobile C-arm. Immediate and continuous control is achieved via 2D anatomy-specific standard views that correspond to a specific C-arm pose relative to the patient positioning, which is currently determined manually, following a trial-and-error approach at the cost of time and radiation dose. The characteristics of the standard views of the knee suggests that the shape information of individual bones could guide an automatic positioning procedure, reducing time and the amount of unnecessary radiation during C-arm positioning. To fully automate the C-arm positioning task during knee surgeries, we propose a complete framework that enables (1) automatic laterality and standard view classification and (2) automatic shape-based pose regression toward the desired standard view based on a single initial X-ray. A suitable shape representation is proposed to incorporate semantic information into the pose regression pipeline. The pipeline is designed to handle two distinct standard views simultaneously. Experiments were conducted to assess the performance of the proposed system on 3528 synthetic and 1386 real X-rays for the a.-p. and lateral standard. The view/laterality classificator resulted in an accuracy of 100\%/98\% on the simulated and 99\%/98\% on the real X-rays. The pose regression performance was $d\theta_{a.-p}=5.8\pm3.3\degree,\,d\theta_{lateral}=3.7\pm2.0\degree$ on the simulated data and $d\theta_{a.-p}=7.4\pm5.0\degree,\,d\theta_{lateral}=8.4\pm5.4\degree$ on the real data outperforming intensity-based pose regression.
Light-weight spatio-temporal graphs for segmentation and ejection fraction prediction in cardiac ultrasound
Jul 06, 2022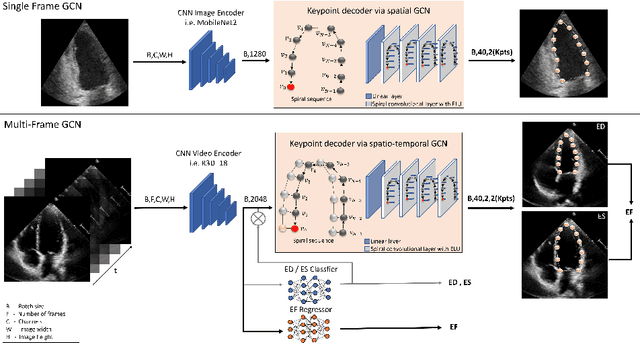
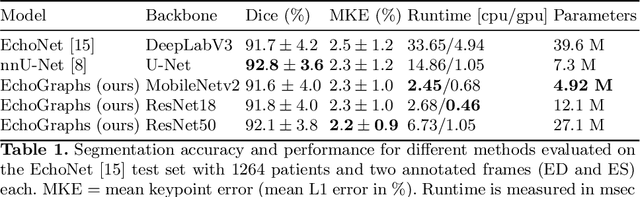
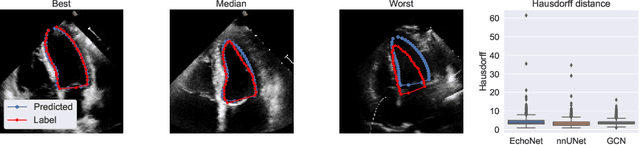
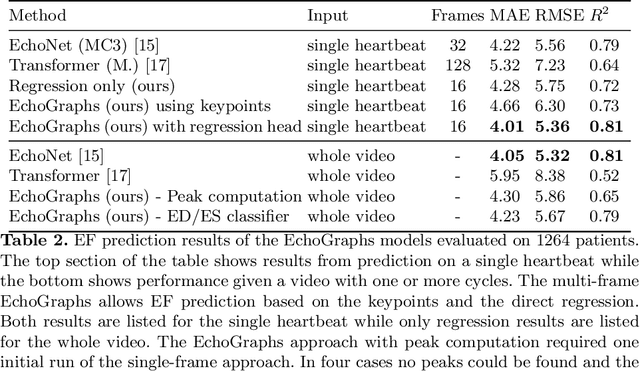
Abstract:Accurate and consistent predictions of echocardiography parameters are important for cardiovascular diagnosis and treatment. In particular, segmentations of the left ventricle can be used to derive ventricular volume, ejection fraction (EF) and other relevant measurements. In this paper we propose a new automated method called EchoGraphs for predicting ejection fraction and segmenting the left ventricle by detecting anatomical keypoints. Models for direct coordinate regression based on Graph Convolutional Networks (GCNs) are used to detect the keypoints. GCNs can learn to represent the cardiac shape based on local appearance of each keypoint, as well as global spatial and temporal structures of all keypoints combined. We evaluate our EchoGraphs model on the EchoNet benchmark dataset. Compared to semantic segmentation, GCNs show accurate segmentation and improvements in robustness and inference runtime. EF is computed simultaneously to segmentations and our method also obtains state-of-the-art ejection fraction estimation. Source code is available online: https://github.com/guybenyosef/EchoGraphs.
Automatic Plane Adjustment of Orthopedic Intra-operative Flat Panel Detector CT-Volumes
Sep 15, 2021



Abstract:Purpose 3D acquisitions are often acquired to assess the result in orthopedic trauma surgery. With a mobile C-Arm system, these acquisitions can be performed intra-operatively. That reduces the number of required revision surgeries. However, due to the operation room setup, the acquisitions typically cannot be performed such that the acquired volumes are aligned to the anatomical regions. Thus, the multiplanar reconstructed (MPR) planes need to be adjusted manually during the review of the volume. In this paper, we present a detailed study of multi-task learning (MTL) regression networks to estimate the parameters of the MPR planes. Approach First, various mathematical descriptions for rotation, including Euler angle, quaternion, and matrix representation, are revised. Then, three different MTL network architectures based on the PoseNet are compared with a single task learning network. Results Using a matrix description rather than the Euler angle description, the accuracy of the regressed normals improves from $7.7^{\circ}$ to $7.3^{\circ}$ in the mean value for single anatomies. The multi-head approach improves the regression of the plane position from $7.4mm$ to $6.1mm$, while the orientation does not benefit from this approach. Conclusions The results show that a multi-head approach can lead to slightly better results than the individual tasks networks. The most important benefit of the MTL approach is that it is a single network for standard plane regression for all body regions with a reduced number of stored parameters.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge