Svein Arne Aase
Low Complexity Point Tracking of the Myocardium in 2D Echocardiography
Mar 13, 2025Abstract:Deep learning methods for point tracking are applicable in 2D echocardiography, but do not yet take advantage of domain specifics that enable extremely fast and efficient configurations. We developed MyoTracker, a low-complexity architecture (0.3M parameters) for point tracking in echocardiography. It builds on the CoTracker2 architecture by simplifying its components and extending the temporal context to provide point predictions for the entire sequence in a single step. We applied MyoTracker to the right ventricular (RV) myocardium in RV-focused recordings and compared the results with those of CoTracker2 and EchoTracker, another specialized point tracking architecture for echocardiography. MyoTracker achieved the lowest average point trajectory error at 2.00 $\pm$ 0.53 mm. Calculating RV Free Wall Strain (RV FWS) using MyoTracker's point predictions resulted in a -0.3$\%$ bias with 95$\%$ limits of agreement from -6.1$\%$ to 5.4$\%$ compared to reference values from commercial software. This range falls within the interobserver variability reported in previous studies. The limits of agreement were wider for both CoTracker2 and EchoTracker, worse than the interobserver variability. At inference, MyoTracker used 67$\%$ less GPU memory than CoTracker2 and 84$\%$ less than EchoTracker on large sequences (100 frames). MyoTracker was 74 times faster during inference than CoTracker2 and 11 times faster than EchoTracker with our setup. Maintaining the entire sequence in the temporal context was the greatest contributor to MyoTracker's accuracy. Slight additional gains can be made by re-enabling iterative refinement, at the cost of longer processing time.
Deep Learning for Multi-Level Detection and Localization of Myocardial Scars Based on Regional Strain Validated on Virtual Patients
Mar 15, 2024Abstract:How well the heart is functioning can be quantified through measurements of myocardial deformation via echocardiography. Clinical assessment of cardiac function is generally focused on global indices of relative shortening, however, territorial, and segmental strain indices have shown to be abnormal in regions of myocardial disease, such as scar. In this work, we propose a single framework to predict myocardial disease substrates at global, territorial, and segmental levels using regional myocardial strain traces as input to a convolutional neural network (CNN)-based classification algorithm. An anatomically meaningful representation of the input data from the clinically standard bullseye representation to a multi-channel 2D image is proposed, to formulate the task as an image classification problem, thus enabling the use of state-of-the-art neural network configurations. A Fully Convolutional Network (FCN) is trained to detect and localize myocardial scar from regional left ventricular (LV) strain patterns. Simulated regional strain data from a controlled dataset of virtual patients with varying degrees and locations of myocardial scar is used for training and validation. The proposed method successfully detects and localizes the scars on 98% of the 5490 left ventricle (LV) segments of the 305 patients in the test set using strain traces only. Due to the sparse existence of scar, only 10% of the LV segments in the virtual patient cohort have scar. Taking the imbalance into account, the class balanced accuracy is calculated as 95%. The performance is reported on global, territorial, and segmental levels. The proposed method proves successful on the strain traces of the virtual cohort and offers the potential to solve the regional myocardial scar detection problem on the strain traces of the real patient cohorts.
* 11 pages, 9 figures and 1 table. Preliminary results of the method was presented as poster in IEEE conference International Ultrasonics Symposium 2022 in Venice, Italy
Cardiac valve event timing in echocardiography using deep learning and triplane recordings
Mar 15, 2024Abstract:Cardiac valve event timing plays a crucial role when conducting clinical measurements using echocardiography. However, established automated approaches are limited by the need of external electrocardiogram sensors, and manual measurements often rely on timing from different cardiac cycles. Recent methods have applied deep learning to cardiac timing, but they have mainly been restricted to only detecting two key time points, namely end-diastole (ED) and end-systole (ES). In this work, we propose a deep learning approach that leverages triplane recordings to enhance detection of valve events in echocardiography. Our method demonstrates improved performance detecting six different events, including valve events conventionally associated with ED and ES. Of all events, we achieve an average absolute frame difference (aFD) of maximum 1.4 frames (29 ms) for start of diastasis, down to 0.6 frames (12 ms) for mitral valve opening when performing a ten-fold cross-validation with test splits on triplane data from 240 patients. On an external independent test consisting of apical long-axis data from 180 other patients, the worst performing event detection had an aFD of 1.8 (30 ms). The proposed approach has the potential to significantly impact clinical practice by enabling more accurate, rapid and comprehensive event detection, leading to improved clinical measurements.
A Data Augmentation Pipeline to Generate Synthetic Labeled Datasets of 3D Echocardiography Images using a GAN
Mar 08, 2024



Abstract:Due to privacy issues and limited amount of publicly available labeled datasets in the domain of medical imaging, we propose an image generation pipeline to synthesize 3D echocardiographic images with corresponding ground truth labels, to alleviate the need for data collection and for laborious and error-prone human labeling of images for subsequent Deep Learning (DL) tasks. The proposed method utilizes detailed anatomical segmentations of the heart as ground truth label sources. This initial dataset is combined with a second dataset made up of real 3D echocardiographic images to train a Generative Adversarial Network (GAN) to synthesize realistic 3D cardiovascular Ultrasound images paired with ground truth labels. To generate the synthetic 3D dataset, the trained GAN uses high resolution anatomical models from Computed Tomography (CT) as input. A qualitative analysis of the synthesized images showed that the main structures of the heart are well delineated and closely follow the labels obtained from the anatomical models. To assess the usability of these synthetic images for DL tasks, segmentation algorithms were trained to delineate the left ventricle, left atrium, and myocardium. A quantitative analysis of the 3D segmentations given by the models trained with the synthetic images indicated the potential use of this GAN approach to generate 3D synthetic data, use the data to train DL models for different clinical tasks, and therefore tackle the problem of scarcity of 3D labeled echocardiography datasets.
Graph Convolutional Neural Networks for Automated Echocardiography View Recognition: A Holistic Approach
Mar 01, 2024Abstract:To facilitate diagnosis on cardiac ultrasound (US), clinical practice has established several standard views of the heart, which serve as reference points for diagnostic measurements and define viewports from which images are acquired. Automatic view recognition involves grouping those images into classes of standard views. Although deep learning techniques have been successful in achieving this, they still struggle with fully verifying the suitability of an image for specific measurements due to factors like the correct location, pose, and potential occlusions of cardiac structures. Our approach goes beyond view classification and incorporates a 3D mesh reconstruction of the heart that enables several more downstream tasks, like segmentation and pose estimation. In this work, we explore learning 3D heart meshes via graph convolutions, using similar techniques to learn 3D meshes in natural images, such as human pose estimation. As the availability of fully annotated 3D images is limited, we generate synthetic US images from 3D meshes by training an adversarial denoising diffusion model. Experiments were conducted on synthetic and clinical cases for view recognition and structure detection. The approach yielded good performance on synthetic images and, despite being exclusively trained on synthetic data, it already showed potential when applied to clinical images. With this proof-of-concept, we aim to demonstrate the benefits of graphs to improve cardiac view recognition that can ultimately lead to better efficiency in cardiac diagnosis.
Doppler Spectrum Classification with CNNs via Heatmap Location Encoding and a Multi-head Output Layer
Nov 08, 2019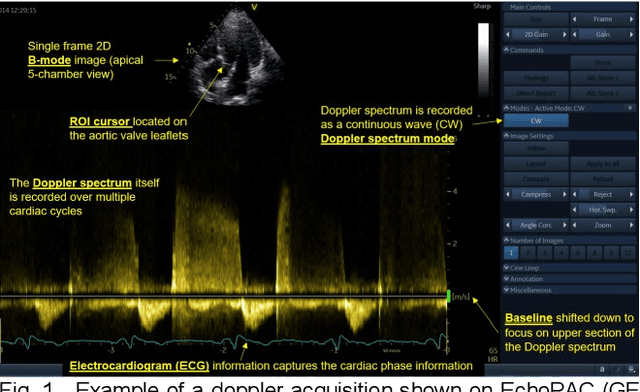
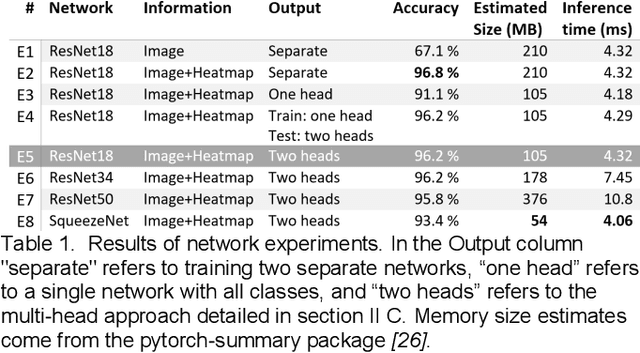
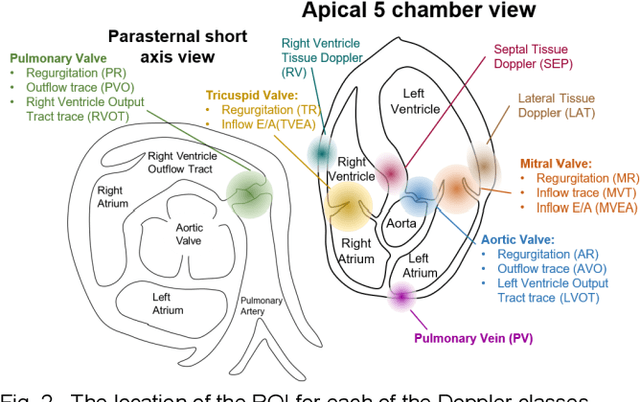

Abstract:Spectral Doppler measurements are an important part of the standard echocardiographic examination. These measurements give important insight into myocardial motion and blood flow providing clinicians with parameters for diagnostic decision making. Many of these measurements can currently be performed automatically with high accuracy, increasing the efficiency of the diagnostic pipeline. However, full automation is not yet available because the user must manually select which measurement should be performed on each image. In this work we develop a convolutional neural network (CNN) to automatically classify cardiac Doppler spectra into measurement classes. We show how the multi-modal information in each spectral Doppler recording can be combined using a meta parameter post-processing mapping scheme and heatmaps to encode coordinate locations. Additionally, we experiment with several state-of-the-art network architectures to examine the tradeoff between accuracy and memory usage for resource-constrained environments. Finally, we propose a confidence metric using the values in the last fully connected layer of the network. We analyze example images that fall outside of our proposed classes to show our confidence metric can prevent many misclassifications. Our algorithm achieves 96% accuracy on a test set drawn from a separate clinical site, indicating that the proposed method is suitable for clinical adoption and enabling a fully automatic pipeline from acquisition to Doppler spectrum measurements.
Automated Left Ventricle Dimension Measurement in 2D Cardiac Ultrasound via an Anatomically Meaningful CNN Approach
Nov 06, 2019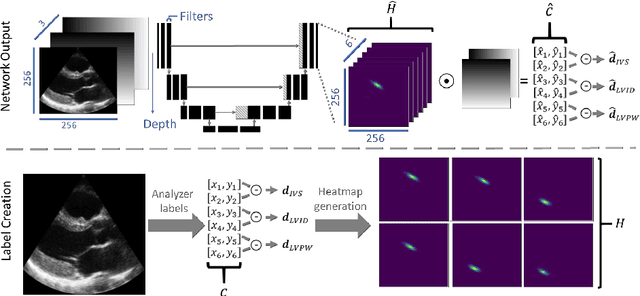
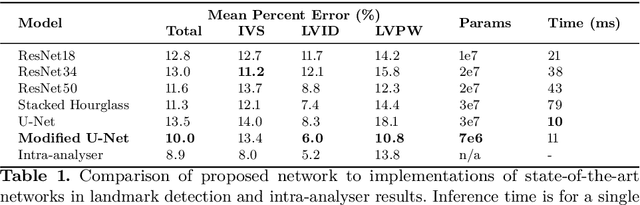
Abstract:Two-dimensional echocardiography (2DE) measurements of left ventricle (LV) dimensions are highly significant markers of several cardiovascular diseases. These measurements are often used in clinical care despite suffering from large variability between observers. This variability is due to the challenging nature of accurately finding the correct temporal and spatial location of measurement endpoints in ultrasound images. These images often contain fuzzy boundaries and varying reflection patterns between frames. In this work, we present a convolutional neural network (CNN) based approach to automate 2DE LV measurements. Treating the problem as a landmark detection problem, we propose a modified U-Net CNN architecture to generate heatmaps of likely coordinate locations. To improve the network performance we use anatomically meaningful heatmaps as labels and train with a multi-component loss function. Our network achieves 13.4%, 6%, and 10.8% mean percent error on intraventricular septum (IVS), LV internal dimension (LVID), and LV posterior wall (LVPW) measurements respectively. The design outperforms other networks and matches or approaches intra-analyser expert error.
* Best paper award at Smart Ultrasound Imaging Workshop (SUSI) MICCAI 2019
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge