Kristin McLeod
A Domain Translation Framework with an Adversarial Denoising Diffusion Model to Generate Synthetic Datasets of Echocardiography Images
Mar 07, 2024Abstract:Currently, medical image domain translation operations show a high demand from researchers and clinicians. Amongst other capabilities, this task allows the generation of new medical images with sufficiently high image quality, making them clinically relevant. Deep Learning (DL) architectures, most specifically deep generative models, are widely used to generate and translate images from one domain to another. The proposed framework relies on an adversarial Denoising Diffusion Model (DDM) to synthesize echocardiography images and perform domain translation. Contrary to Generative Adversarial Networks (GANs), DDMs are able to generate high quality image samples with a large diversity. If a DDM is combined with a GAN, this ability to generate new data is completed at an even faster sampling time. In this work we trained an adversarial DDM combined with a GAN to learn the reverse denoising process, relying on a guide image, making sure relevant anatomical structures of each echocardiography image were kept and represented on the generated image samples. For several domain translation operations, the results verified that such generative model was able to synthesize high quality image samples: MSE: 11.50 +/- 3.69, PSNR (dB): 30.48 +/- 0.09, SSIM: 0.47 +/- 0.03. The proposed method showed high generalization ability, introducing a framework to create echocardiography images suitable to be used for clinical research purposes.
Doppler Spectrum Classification with CNNs via Heatmap Location Encoding and a Multi-head Output Layer
Nov 08, 2019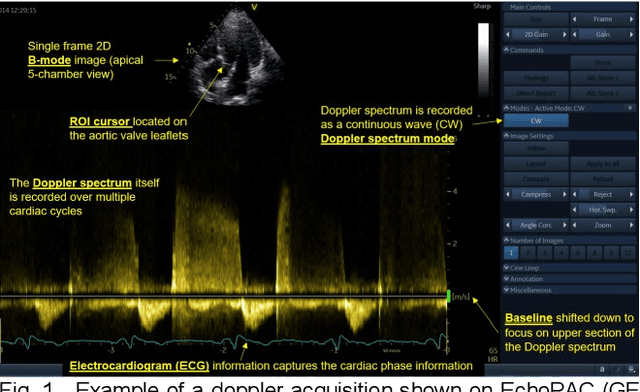
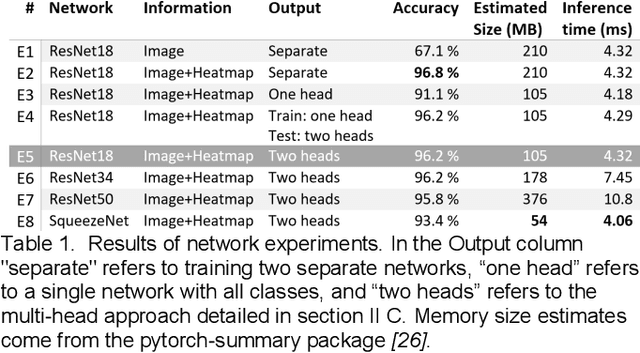
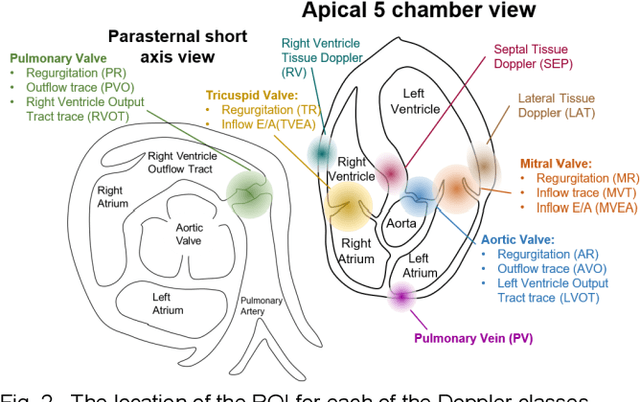

Abstract:Spectral Doppler measurements are an important part of the standard echocardiographic examination. These measurements give important insight into myocardial motion and blood flow providing clinicians with parameters for diagnostic decision making. Many of these measurements can currently be performed automatically with high accuracy, increasing the efficiency of the diagnostic pipeline. However, full automation is not yet available because the user must manually select which measurement should be performed on each image. In this work we develop a convolutional neural network (CNN) to automatically classify cardiac Doppler spectra into measurement classes. We show how the multi-modal information in each spectral Doppler recording can be combined using a meta parameter post-processing mapping scheme and heatmaps to encode coordinate locations. Additionally, we experiment with several state-of-the-art network architectures to examine the tradeoff between accuracy and memory usage for resource-constrained environments. Finally, we propose a confidence metric using the values in the last fully connected layer of the network. We analyze example images that fall outside of our proposed classes to show our confidence metric can prevent many misclassifications. Our algorithm achieves 96% accuracy on a test set drawn from a separate clinical site, indicating that the proposed method is suitable for clinical adoption and enabling a fully automatic pipeline from acquisition to Doppler spectrum measurements.
Automated Left Ventricle Dimension Measurement in 2D Cardiac Ultrasound via an Anatomically Meaningful CNN Approach
Nov 06, 2019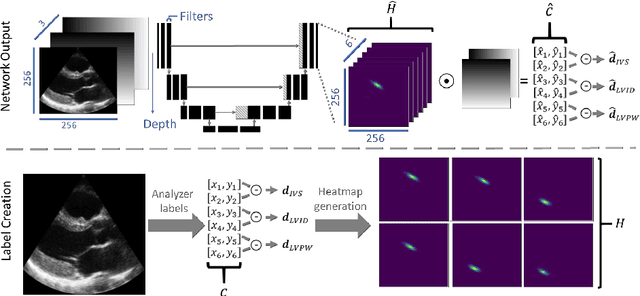
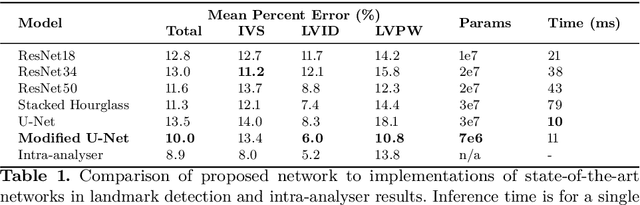
Abstract:Two-dimensional echocardiography (2DE) measurements of left ventricle (LV) dimensions are highly significant markers of several cardiovascular diseases. These measurements are often used in clinical care despite suffering from large variability between observers. This variability is due to the challenging nature of accurately finding the correct temporal and spatial location of measurement endpoints in ultrasound images. These images often contain fuzzy boundaries and varying reflection patterns between frames. In this work, we present a convolutional neural network (CNN) based approach to automate 2DE LV measurements. Treating the problem as a landmark detection problem, we propose a modified U-Net CNN architecture to generate heatmaps of likely coordinate locations. To improve the network performance we use anatomically meaningful heatmaps as labels and train with a multi-component loss function. Our network achieves 13.4%, 6%, and 10.8% mean percent error on intraventricular septum (IVS), LV internal dimension (LVID), and LV posterior wall (LVPW) measurements respectively. The design outperforms other networks and matches or approaches intra-analyser expert error.
* Best paper award at Smart Ultrasound Imaging Workshop (SUSI) MICCAI 2019
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge