Andreas Østvik
Low Complexity Point Tracking of the Myocardium in 2D Echocardiography
Mar 13, 2025Abstract:Deep learning methods for point tracking are applicable in 2D echocardiography, but do not yet take advantage of domain specifics that enable extremely fast and efficient configurations. We developed MyoTracker, a low-complexity architecture (0.3M parameters) for point tracking in echocardiography. It builds on the CoTracker2 architecture by simplifying its components and extending the temporal context to provide point predictions for the entire sequence in a single step. We applied MyoTracker to the right ventricular (RV) myocardium in RV-focused recordings and compared the results with those of CoTracker2 and EchoTracker, another specialized point tracking architecture for echocardiography. MyoTracker achieved the lowest average point trajectory error at 2.00 $\pm$ 0.53 mm. Calculating RV Free Wall Strain (RV FWS) using MyoTracker's point predictions resulted in a -0.3$\%$ bias with 95$\%$ limits of agreement from -6.1$\%$ to 5.4$\%$ compared to reference values from commercial software. This range falls within the interobserver variability reported in previous studies. The limits of agreement were wider for both CoTracker2 and EchoTracker, worse than the interobserver variability. At inference, MyoTracker used 67$\%$ less GPU memory than CoTracker2 and 84$\%$ less than EchoTracker on large sequences (100 frames). MyoTracker was 74 times faster during inference than CoTracker2 and 11 times faster than EchoTracker with our setup. Maintaining the entire sequence in the temporal context was the greatest contributor to MyoTracker's accuracy. Slight additional gains can be made by re-enabling iterative refinement, at the cost of longer processing time.
Regional quality estimation for echocardiography using deep learning
Aug 01, 2024Abstract:Automatic estimation of cardiac ultrasound image quality can be beneficial for guiding operators and ensuring the accuracy of clinical measurements. Previous work often fails to distinguish the view correctness of the echocardiogram from the image quality. Additionally, previous studies only provide a global image quality value, which limits their practical utility. In this work, we developed and compared three methods to estimate image quality: 1) classic pixel-based metrics like the generalized contrast-to-noise ratio (gCNR) on myocardial segments as region of interest and left ventricle lumen as background, obtained using a U-Net segmentation 2) local image coherence derived from a U-Net model that predicts coherence from B-Mode images 3) a deep convolutional network that predicts the quality of each region directly in an end-to-end fashion. We evaluate each method against manual regional image quality annotations by three experienced cardiologists. The results indicate poor performance of the gCNR metric, with Spearman correlation to the annotations of \r{ho} = 0.24. The end-to-end learning model obtains the best result, \r{ho} = 0.69, comparable to the inter-observer correlation, \r{ho} = 0.63. Finally, the coherence-based method, with \r{ho} = 0.58, outperformed the classical metrics and is more generic than the end-to-end approach.
EchoTracker: Advancing Myocardial Point Tracking in Echocardiography
May 14, 2024Abstract:Tissue tracking in echocardiography is challenging due to the complex cardiac motion and the inherent nature of ultrasound acquisitions. Although optical flow methods are considered state-of-the-art (SOTA), they struggle with long-range tracking, noise occlusions, and drift throughout the cardiac cycle. Recently, novel learning-based point tracking techniques have been introduced to tackle some of these issues. In this paper, we build upon these techniques and introduce EchoTracker, a two-fold coarse-to-fine model that facilitates the tracking of queried points on a tissue surface across ultrasound image sequences. The architecture contains a preliminary coarse initialization of the trajectories, followed by reinforcement iterations based on fine-grained appearance changes. It is efficient, light, and can run on mid-range GPUs. Experiments demonstrate that the model outperforms SOTA methods, with an average position accuracy of 67% and a median trajectory error of 2.86 pixels. Furthermore, we show a relative improvement of 25% when using our model to calculate the global longitudinal strain (GLS) in a clinical test-retest dataset compared to other methods. This implies that learning-based point tracking can potentially improve performance and yield a higher diagnostic and prognostic value for clinical measurements than current techniques. Our source code is available at: https://github.com/riponazad/echotracker/.
Cardiac valve event timing in echocardiography using deep learning and triplane recordings
Mar 15, 2024Abstract:Cardiac valve event timing plays a crucial role when conducting clinical measurements using echocardiography. However, established automated approaches are limited by the need of external electrocardiogram sensors, and manual measurements often rely on timing from different cardiac cycles. Recent methods have applied deep learning to cardiac timing, but they have mainly been restricted to only detecting two key time points, namely end-diastole (ED) and end-systole (ES). In this work, we propose a deep learning approach that leverages triplane recordings to enhance detection of valve events in echocardiography. Our method demonstrates improved performance detecting six different events, including valve events conventionally associated with ED and ES. Of all events, we achieve an average absolute frame difference (aFD) of maximum 1.4 frames (29 ms) for start of diastasis, down to 0.6 frames (12 ms) for mitral valve opening when performing a ten-fold cross-validation with test splits on triplane data from 240 patients. On an external independent test consisting of apical long-axis data from 180 other patients, the worst performing event detection had an aFD of 1.8 (30 ms). The proposed approach has the potential to significantly impact clinical practice by enabling more accurate, rapid and comprehensive event detection, leading to improved clinical measurements.
LU-Net: a multi-task network to improve the robustness of segmentation of left ventriclular structures by deep learning in 2D echocardiography
Apr 04, 2020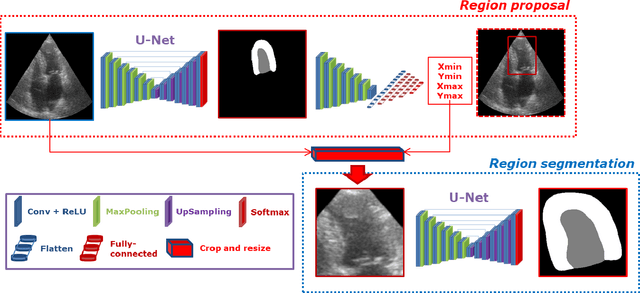
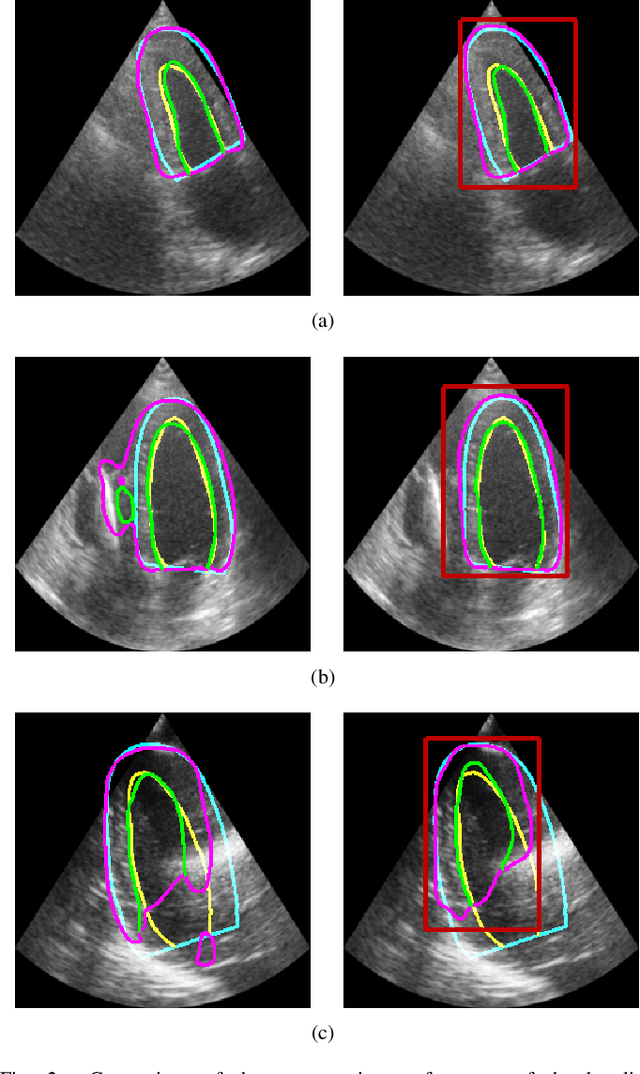
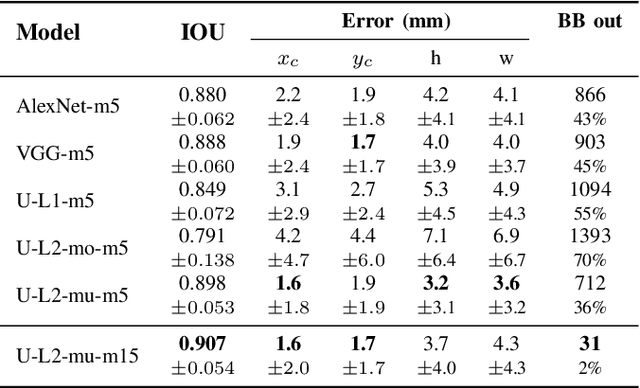
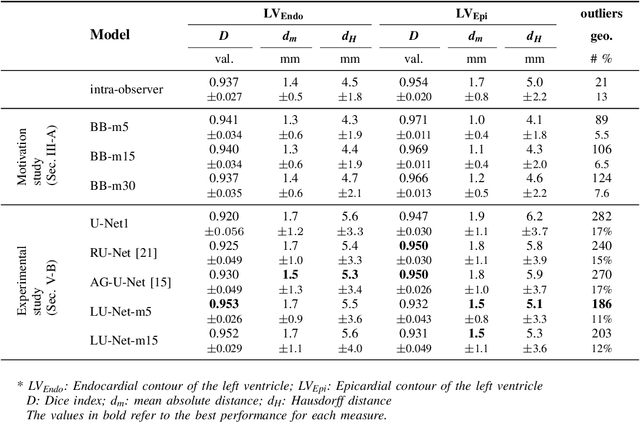
Abstract:Segmentation of cardiac structures is one of the fundamental steps to estimate volumetric indices of the heart. This step is still performed semi-automatically in clinical routine, and is thus prone to inter- and intra-observer variability. Recent studies have shown that deep learning has the potential to perform fully automatic segmentation. However, the current best solutions still suffer from a lack of robustness. In this work, we introduce an end-to-end multi-task network designed to improve the overall accuracy of cardiac segmentation while enhancing the estimation of clinical indices and reducing the number of outliers. Results obtained on a large open access dataset show that our method outperforms the current best performing deep learning solution and achieved an overall segmentation accuracy lower than the intra-observer variability for the epicardial border (i.e. on average a mean absolute error of 1.5mm and a Hausdorff distance of 5.1mm) with 11% of outliers. Moreover, we demonstrate that our method can closely reproduce the expert analysis for the end-diastolic and end-systolic left ventricular volumes, with a mean correlation of 0.96 and a mean absolute error of 7.6ml. Concerning the ejection fraction of the left ventricle, results are more contrasted with a mean correlation coefficient of 0.83 and an absolute mean error of 5.0%, producing scores that are slightly below the intra-observer margin. Based on this observation, areas for improvement are suggested.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge