Thomas Grenier
Enhanced segmentation of femoral bone metastasis in CT scans of patients using synthetic data generation with 3D diffusion models
Sep 17, 2024



Abstract:Purpose: Bone metastasis have a major impact on the quality of life of patients and they are diverse in terms of size and location, making their segmentation complex. Manual segmentation is time-consuming, and expert segmentations are subject to operator variability, which makes obtaining accurate and reproducible segmentations of bone metastasis on CT-scans a challenging yet important task to achieve. Materials and Methods: Deep learning methods tackle segmentation tasks efficiently but require large datasets along with expert manual segmentations to generalize on new images. We propose an automated data synthesis pipeline using 3D Denoising Diffusion Probabilistic Models (DDPM) to enchance the segmentation of femoral metastasis from CT-scan volumes of patients. We used 29 existing lesions along with 26 healthy femurs to create new realistic synthetic metastatic images, and trained a DDPM to improve the diversity and realism of the simulated volumes. We also investigated the operator variability on manual segmentation. Results: We created 5675 new volumes, then trained 3D U-Net segmentation models on real and synthetic data to compare segmentation performance, and we evaluated the performance of the models depending on the amount of synthetic data used in training. Conclusion: Our results showed that segmentation models trained with synthetic data outperformed those trained on real volumes only, and that those models perform especially well when considering operator variability.
LU-Net: a multi-task network to improve the robustness of segmentation of left ventriclular structures by deep learning in 2D echocardiography
Apr 04, 2020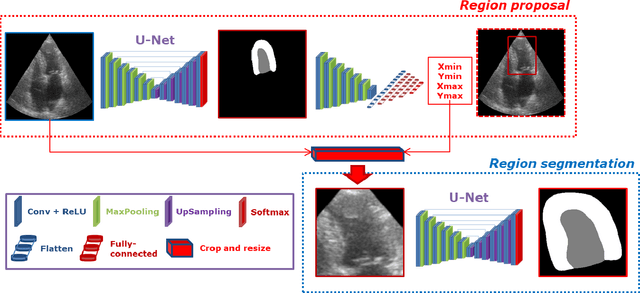
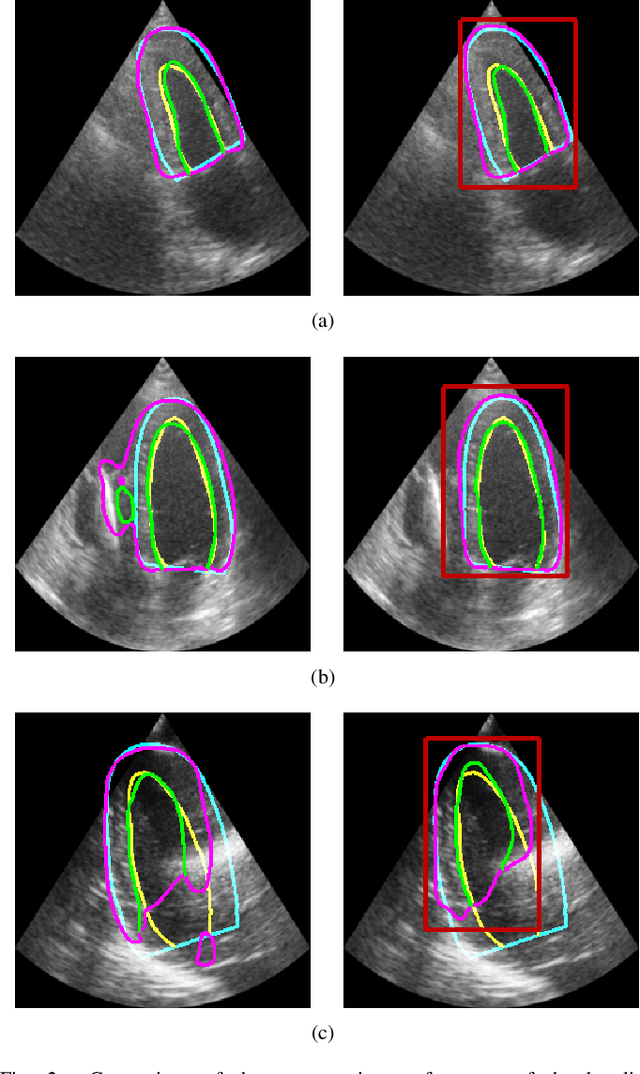
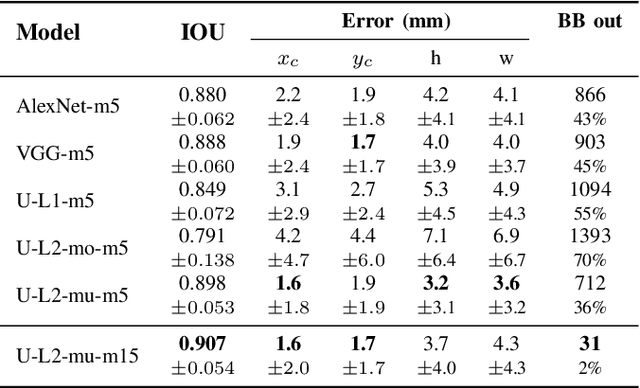
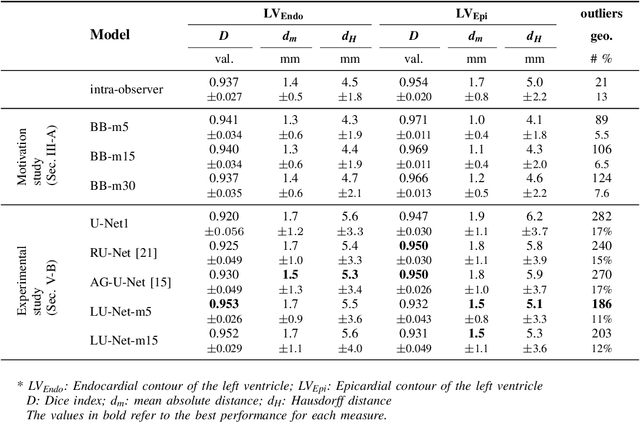
Abstract:Segmentation of cardiac structures is one of the fundamental steps to estimate volumetric indices of the heart. This step is still performed semi-automatically in clinical routine, and is thus prone to inter- and intra-observer variability. Recent studies have shown that deep learning has the potential to perform fully automatic segmentation. However, the current best solutions still suffer from a lack of robustness. In this work, we introduce an end-to-end multi-task network designed to improve the overall accuracy of cardiac segmentation while enhancing the estimation of clinical indices and reducing the number of outliers. Results obtained on a large open access dataset show that our method outperforms the current best performing deep learning solution and achieved an overall segmentation accuracy lower than the intra-observer variability for the epicardial border (i.e. on average a mean absolute error of 1.5mm and a Hausdorff distance of 5.1mm) with 11% of outliers. Moreover, we demonstrate that our method can closely reproduce the expert analysis for the end-diastolic and end-systolic left ventricular volumes, with a mean correlation of 0.96 and a mean absolute error of 7.6ml. Concerning the ejection fraction of the left ventricle, results are more contrasted with a mean correlation coefficient of 0.83 and an absolute mean error of 5.0%, producing scores that are slightly below the intra-observer margin. Based on this observation, areas for improvement are suggested.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge