Lasse Løvstakken
Lightweight image segmentation for echocardiography
Sep 03, 2025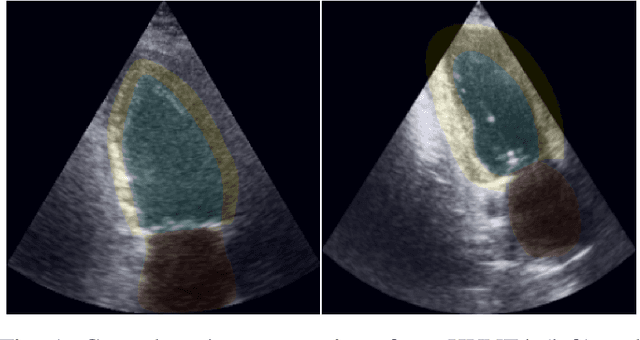
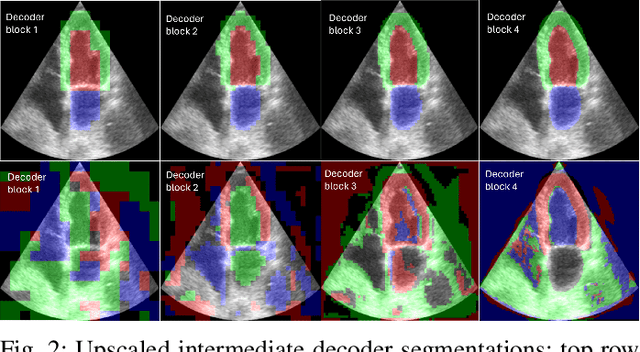
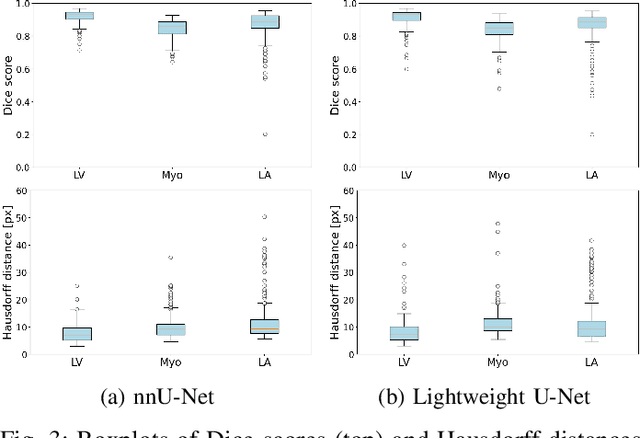
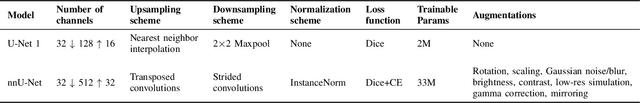
Abstract:Accurate segmentation of the left ventricle in echocardiography can enable fully automatic extraction of clinical measurements such as volumes and ejection fraction. While models configured by nnU-Net perform well, they are large and slow, thus limiting real-time use. We identified the most effective components of nnU-Net for cardiac segmentation through an ablation study, incrementally evaluating data augmentation schemes, architectural modifications, loss functions, and post-processing techniques. Our analysis revealed that simple affine augmentations and deep supervision drive performance, while complex augmentations and large model capacity offer diminishing returns. Based on these insights, we developed a lightweight U-Net (2M vs 33M parameters) that achieves statistically equivalent performance to nnU-Net on CAMUS (N=500) with Dice scores of 0.93/0.85/0.89 vs 0.93/0.86/0.89 for LV/MYO/LA ($p>0.05$), while being 16 times smaller and 4 times faster (1.35ms vs 5.40ms per frame) than the default nnU-Net configuration. Cross-dataset evaluation on an internal dataset (N=311) confirms comparable generalization.
Generative augmentations for improved cardiac ultrasound segmentation using diffusion models
Feb 27, 2025Abstract:One of the main challenges in current research on segmentation in cardiac ultrasound is the lack of large and varied labeled datasets and the differences in annotation conventions between datasets. This makes it difficult to design robust segmentation models that generalize well to external datasets. This work utilizes diffusion models to create generative augmentations that can significantly improve diversity of the dataset and thus the generalisability of segmentation models without the need for more annotated data. The augmentations are applied in addition to regular augmentations. A visual test survey showed that experts cannot clearly distinguish between real and fully generated images. Using the proposed generative augmentations, segmentation robustness was increased when training on an internal dataset and testing on an external dataset with an improvement of over 20 millimeters in Hausdorff distance. Additionally, the limits of agreement for automatic ejection fraction estimation improved by up to 20% of absolute ejection fraction value on out of distribution cases. These improvements come exclusively from the increased variation of the training data using the generative augmentations, without modifying the underlying machine learning model. The augmentation tool is available as an open source Python library at https://github.com/GillesVanDeVyver/EchoGAINS.
Regional quality estimation for echocardiography using deep learning
Aug 01, 2024Abstract:Automatic estimation of cardiac ultrasound image quality can be beneficial for guiding operators and ensuring the accuracy of clinical measurements. Previous work often fails to distinguish the view correctness of the echocardiogram from the image quality. Additionally, previous studies only provide a global image quality value, which limits their practical utility. In this work, we developed and compared three methods to estimate image quality: 1) classic pixel-based metrics like the generalized contrast-to-noise ratio (gCNR) on myocardial segments as region of interest and left ventricle lumen as background, obtained using a U-Net segmentation 2) local image coherence derived from a U-Net model that predicts coherence from B-Mode images 3) a deep convolutional network that predicts the quality of each region directly in an end-to-end fashion. We evaluate each method against manual regional image quality annotations by three experienced cardiologists. The results indicate poor performance of the gCNR metric, with Spearman correlation to the annotations of \r{ho} = 0.24. The end-to-end learning model obtains the best result, \r{ho} = 0.69, comparable to the inter-observer correlation, \r{ho} = 0.63. Finally, the coherence-based method, with \r{ho} = 0.58, outperformed the classical metrics and is more generic than the end-to-end approach.
Towards Robust Cardiac Segmentation using Graph Convolutional Networks
Oct 02, 2023



Abstract:Fully automatic cardiac segmentation can be a fast and reproducible method to extract clinical measurements from an echocardiography examination. The U-Net architecture is the current state-of-the-art deep learning architecture for medical segmentation and can segment cardiac structures in real-time with average errors comparable to inter-observer variability. However, this architecture still generates large outliers that are often anatomically incorrect. This work uses the concept of graph convolutional neural networks that predict the contour points of the structures of interest instead of labeling each pixel. We propose a graph architecture that uses two convolutional rings based on cardiac anatomy and show that this eliminates anatomical incorrect multi-structure segmentations on the publicly available CAMUS dataset. Additionally, this work contributes with an ablation study on the graph convolutional architecture and an evaluation of clinical measurements on the clinical HUNT4 dataset. Finally, we propose to use the inter-model agreement of the U-Net and the graph network as a predictor of both the input and segmentation quality. We show this predictor can detect out-of-distribution and unsuitable input images in real-time. Source code is available online: https://github.com/gillesvntnu/GCN_multistructure
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge