Sarath Swaminathan
Larimar: Large Language Models with Episodic Memory Control
Mar 18, 2024Abstract:Efficient and accurate updating of knowledge stored in Large Language Models (LLMs) is one of the most pressing research challenges today. This paper presents Larimar - a novel, brain-inspired architecture for enhancing LLMs with a distributed episodic memory. Larimar's memory allows for dynamic, one-shot updates of knowledge without the need for computationally expensive re-training or fine-tuning. Experimental results on multiple fact editing benchmarks demonstrate that Larimar attains accuracy comparable to most competitive baselines, even in the challenging sequential editing setup, but also excels in speed - yielding speed-ups of 4-10x depending on the base LLM - as well as flexibility due to the proposed architecture being simple, LLM-agnostic, and hence general. We further provide mechanisms for selective fact forgetting and input context length generalization with Larimar and show their effectiveness.
Language models in molecular discovery
Sep 28, 2023Abstract:The success of language models, especially transformer-based architectures, has trickled into other domains giving rise to "scientific language models" that operate on small molecules, proteins or polymers. In chemistry, language models contribute to accelerating the molecule discovery cycle as evidenced by promising recent findings in early-stage drug discovery. Here, we review the role of language models in molecular discovery, underlining their strength in de novo drug design, property prediction and reaction chemistry. We highlight valuable open-source software assets thus lowering the entry barrier to the field of scientific language modeling. Last, we sketch a vision for future molecular design that combines a chatbot interface with access to computational chemistry tools. Our contribution serves as a valuable resource for researchers, chemists, and AI enthusiasts interested in understanding how language models can and will be used to accelerate chemical discovery.
Data Infrastructure and Approaches for Ontology-Based Drug Repurposing
Jul 12, 2018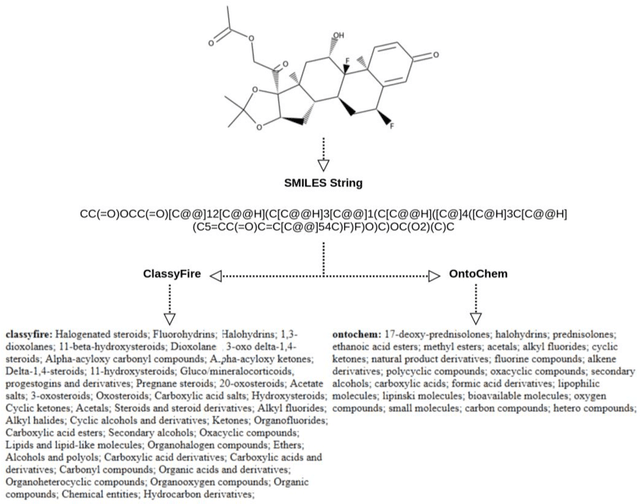
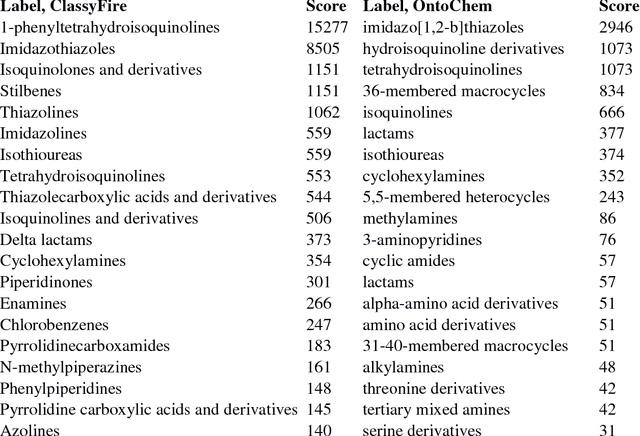
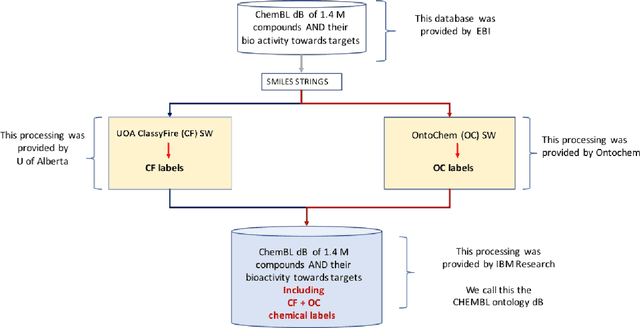

Abstract:We report development of a data infrastructure for drug repurposing that takes advantage of two currently available chemical ontologies. The data infrastructure includes a database of compound- target associations augmented with molecular ontological labels. It also contains two computational tools for prediction of new associations. We describe two drug-repurposing systems: one, Nascent Ontological Information Retrieval for Drug Repurposing (NOIR-DR), based on an information retrieval strategy, and another, based on non-negative matrix factorization together with compound similarity, that was inspired by recommender systems. We report the performance of both tools on a drug-repurposing task.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge