Sarah Lippe
University of Montreal, Canada, CHU Sainte-Justine Research Center, Montreal, Canada
Conditional canonical correlation estimation based on covariates with random forests
Nov 23, 2020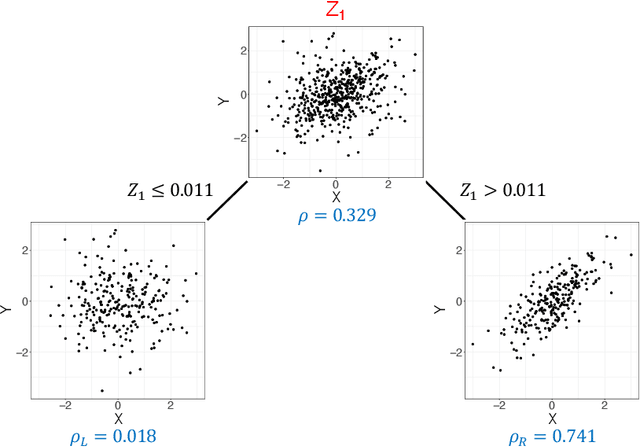
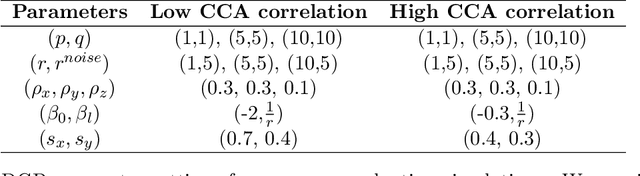
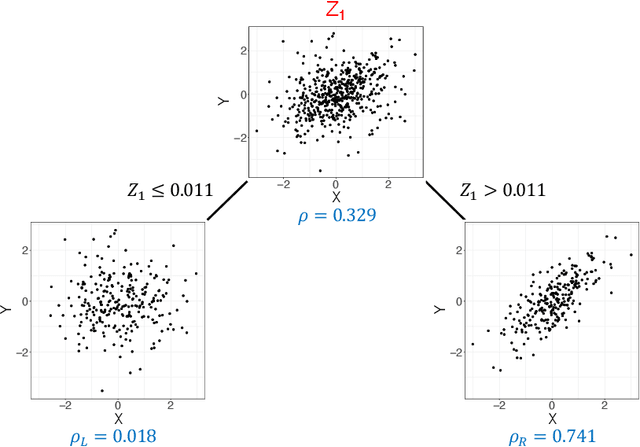
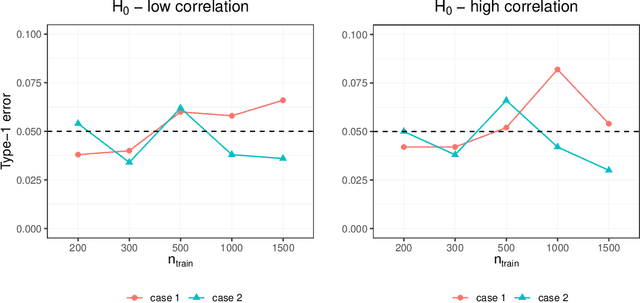
Abstract:Investigating the relationships between two sets of variables helps to understand their interactions and can be done with canonical correlation analysis (CCA). However, the correlation between the two sets can sometimes depend on a third set of covariates, often subject-related ones such as age, gender, or other clinical measures. In this case, applying CCA to the whole population is not optimal and methods to estimate conditional CCA, given the covariates, can be useful. We propose a new method called Random Forest with Canonical Correlation Analysis (RFCCA) to estimate the conditional canonical correlations between two sets of variables given subject-related covariates. The individual trees in the forest are built with a splitting rule specifically designed to partition the data to maximize the canonical correlation heterogeneity between child nodes. We also propose a significance test to detect the global effect of the covariates on the relationship between two sets of variables. The performance of the proposed method and the global significance test is evaluated through simulation studies that show it provides accurate canonical correlation estimations and well-controlled Type-1 error. We also show an application of the proposed method with EEG data.
Dilatation of Lateral Ventricles with Brain Volumes in Infants with 3D Transfontanelle US
Jun 06, 2018

Abstract:Ultrasound (US) can be used to assess brain development in newborns, as MRI is challenging due to immobilization issues, and may require sedation. Dilatation of the lateral ventricles in the brain is a risk factor for poorer neurodevelopment outcomes in infants. Hence, 3D US has the ability to assess the volume of the lateral ventricles similar to clinically standard MRI, but manual segmentation is time consuming. The objective of this study is to develop an approach quantifying the ratio of lateral ventricular dilatation with respect to total brain volume using 3D US, which can assess the severity of macrocephaly. Automatic segmentation of the lateral ventricles is achieved with a multi-atlas deformable registration approach using locally linear correlation metrics for US-MRI fusion, followed by a refinement step using deformable mesh models. Total brain volume is estimated using a 3D ellipsoid modeling approach. Validation was performed on a cohort of 12 infants, ranging from 2 to 8.5 months old, where 3D US and MRI were used to compare brain volumes and segmented lateral ventricles. Automatically extracted volumes from 3D US show a high correlation and no statistically significant difference when compared to ground truth measurements. Differences in volume ratios was 6.0 +/- 4.8% compared to MRI, while lateral ventricular segmentation yielded a mean Dice coefficient of 70.8 +/- 3.6% and a mean absolute distance (MAD) of 0.88 +/- 0.2mm, demonstrating the clinical benefit of this tool in paediatric ultrasound.
Prior-based Coregistration and Cosegmentation
Jul 22, 2016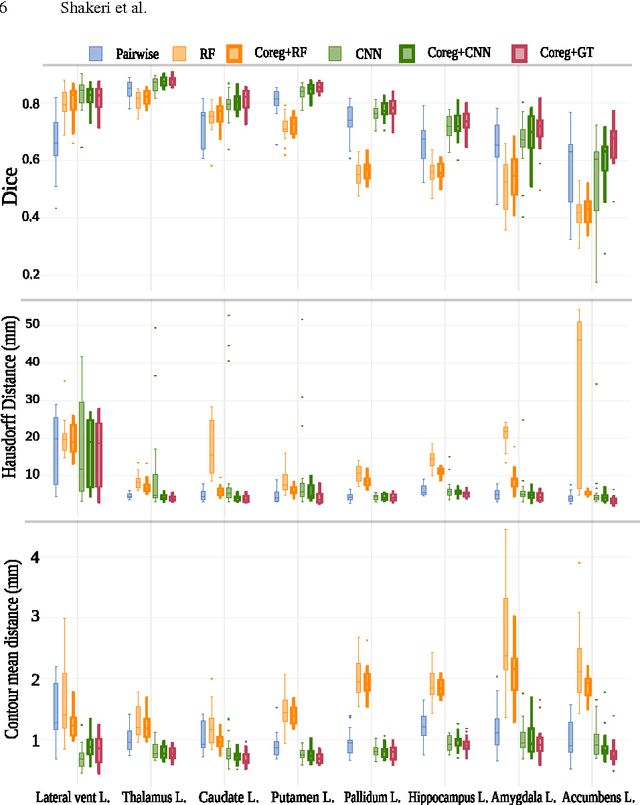
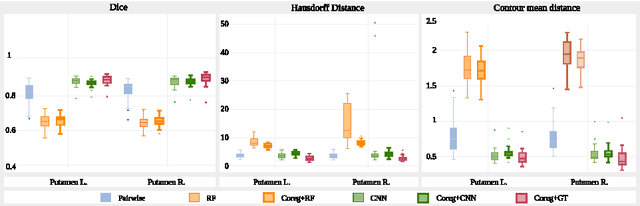
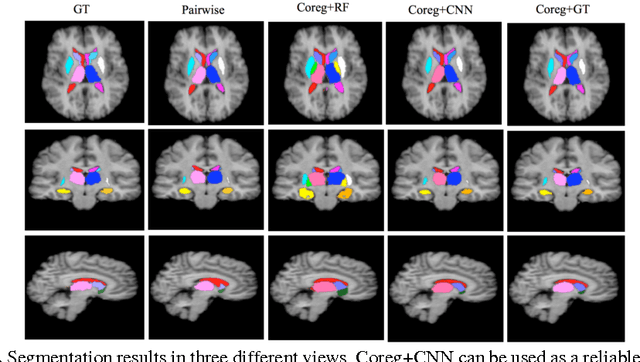
Abstract:We propose a modular and scalable framework for dense coregistration and cosegmentation with two key characteristics: first, we substitute ground truth data with the semantic map output of a classifier; second, we combine this output with population deformable registration to improve both alignment and segmentation. Our approach deforms all volumes towards consensus, taking into account image similarities and label consistency. Our pipeline can incorporate any classifier and similarity metric. Results on two datasets, containing annotations of challenging brain structures, demonstrate the potential of our method.
* The first two authors contributed equally
Sub-cortical brain structure segmentation using F-CNN's
Feb 05, 2016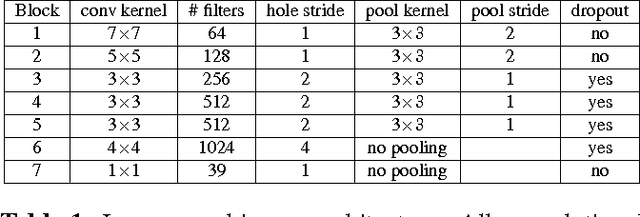
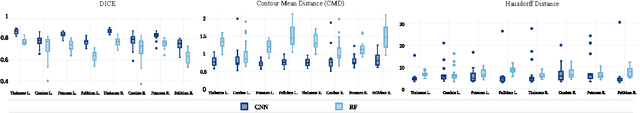
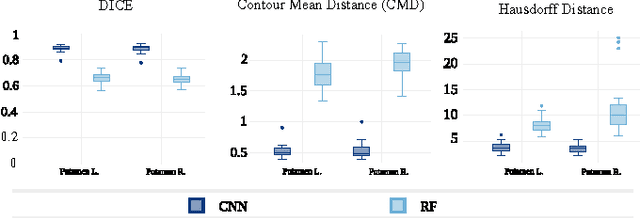
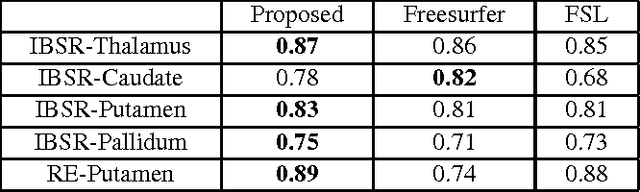
Abstract:In this paper we propose a deep learning approach for segmenting sub-cortical structures of the human brain in Magnetic Resonance (MR) image data. We draw inspiration from a state-of-the-art Fully-Convolutional Neural Network (F-CNN) architecture for semantic segmentation of objects in natural images, and adapt it to our task. Unlike previous CNN-based methods that operate on image patches, our model is applied on a full blown 2D image, without any alignment or registration steps at testing time. We further improve segmentation results by interpreting the CNN output as potentials of a Markov Random Field (MRF), whose topology corresponds to a volumetric grid. Alpha-expansion is used to perform approximate inference imposing spatial volumetric homogeneity to the CNN priors. We compare the performance of the proposed pipeline with a similar system using Random Forest-based priors, as well as state-of-art segmentation algorithms, and show promising results on two different brain MRI datasets.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge