Roberto Annunziata
Jointly Aligning Millions of Images with Deep Penalised Reconstruction Congealing
Aug 12, 2019
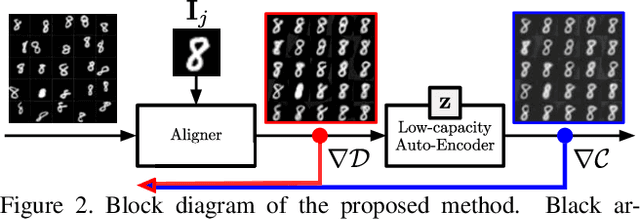
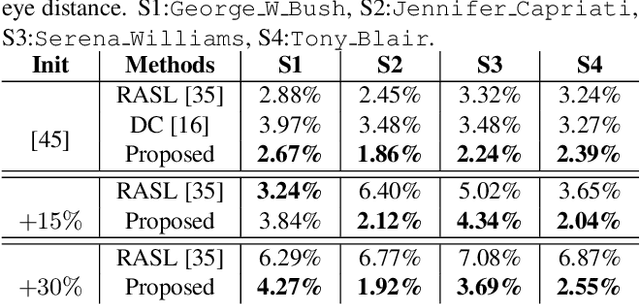
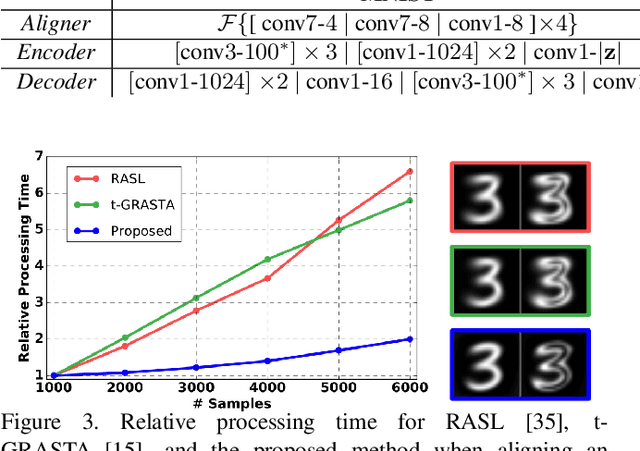
Abstract:Extrapolating fine-grained pixel-level correspondences in a fully unsupervised manner from a large set of misaligned images can benefit several computer vision and graphics problems, e.g. co-segmentation, super-resolution, image edit propagation, structure-from-motion, and 3D reconstruction. Several joint image alignment and congealing techniques have been proposed to tackle this problem, but robustness to initialisation, ability to scale to large datasets, and alignment accuracy seem to hamper their wide applicability. To overcome these limitations, we propose an unsupervised joint alignment method leveraging a densely fused spatial transformer network to estimate the warping parameters for each image and a low-capacity auto-encoder whose reconstruction error is used as an auxiliary measure of joint alignment. Experimental results on digits from multiple versions of MNIST (i.e., original, perturbed, affNIST and infiMNIST) and faces from LFW, show that our approach is capable of aligning millions of images with high accuracy and robustness to different levels and types of perturbation. Moreover, qualitative and quantitative results suggest that the proposed method outperforms state-of-the-art approaches both in terms of alignment quality and robustness to initialisation.
DeSTNet: Densely Fused Spatial Transformer Networks
Jul 16, 2018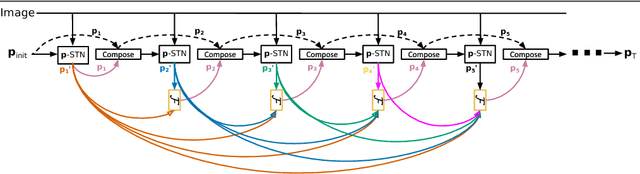
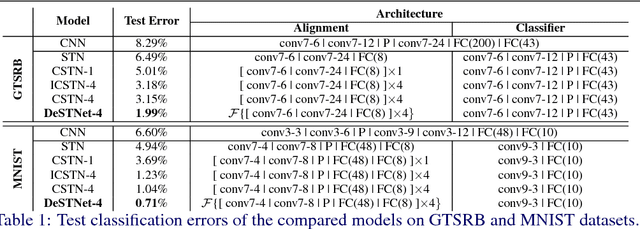

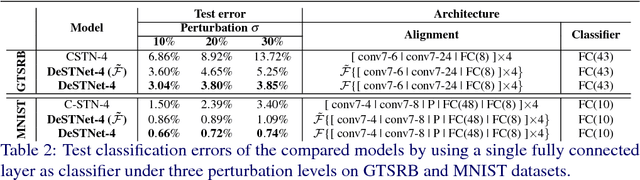
Abstract:Modern Convolutional Neural Networks (CNN) are extremely powerful on a range of computer vision tasks. However, their performance may degrade when the data is characterised by large intra-class variability caused by spatial transformations. The Spatial Transformer Network (STN) is currently the method of choice for providing CNNs the ability to remove those transformations and improve performance in an end-to-end learning framework. In this paper, we propose Densely Fused Spatial Transformer Network (DeSTNet), which, to our best knowledge, is the first dense fusion pattern for combining multiple STNs. Specifically, we show how changing the connectivity pattern of multiple STNs from sequential to dense leads to more powerful alignment modules. Extensive experiments on three benchmarks namely, MNIST, GTSRB, and IDocDB show that the proposed technique outperforms related state-of-the-art methods (i.e., STNs and CSTNs) both in terms of accuracy and robustness.
Joint registration and synthesis using a probabilistic model for alignment of MRI and histological sections
Jan 16, 2018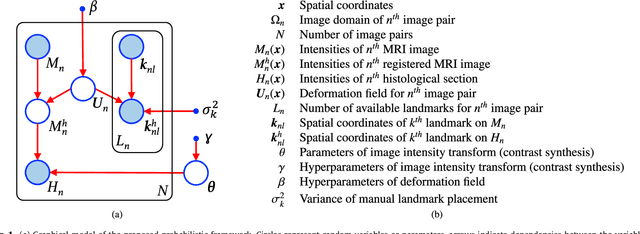
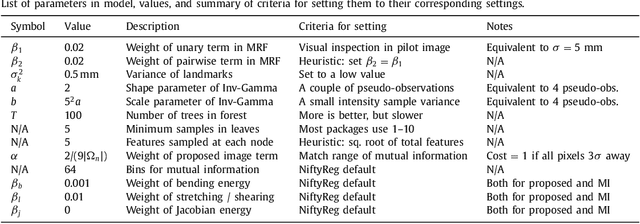
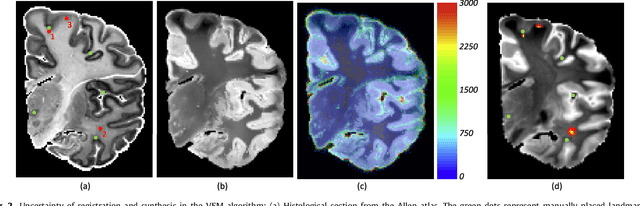
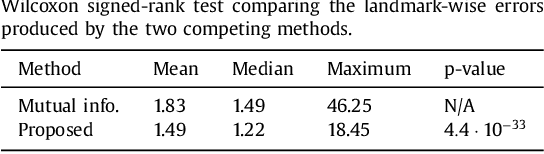
Abstract:Nonlinear registration of 2D histological sections with corresponding slices of MRI data is a critical step of 3D histology reconstruction. This task is difficult due to the large differences in image contrast and resolution, as well as the complex nonrigid distortions produced when sectioning the sample and mounting it on the glass slide. It has been shown in brain MRI registration that better spatial alignment across modalities can be obtained by synthesizing one modality from the other and then using intra-modality registration metrics, rather than by using mutual information (MI) as metric. However, such an approach typically requires a database of aligned images from the two modalities, which is very difficult to obtain for histology/MRI. Here, we overcome this limitation with a probabilistic method that simultaneously solves for registration and synthesis directly on the target images, without any training data. In our model, the MRI slice is assumed to be a contrast-warped, spatially deformed version of the histological section. We use approximate Bayesian inference to iteratively refine the probabilistic estimate of the synthesis and the registration, while accounting for each other's uncertainty. Moreover, manually placed landmarks can be seamlessly integrated in the framework for increased performance. Experiments on a synthetic dataset show that, compared with MI, the proposed method makes it possible to use a much more flexible deformation model in the registration to improve its accuracy, without compromising robustness. Moreover, our framework also exploits information in manually placed landmarks more efficiently than MI, since landmarks inform both synthesis and registration - as opposed to registration alone. Finally, we show qualitative results on the public Allen atlas, in which the proposed method provides a clear improvement over MI based registration.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge