Ramsey D. Badawi
Anatomically and Metabolically Informed Diffusion for Unified Denoising and Segmentation in Low-Count PET Imaging
Mar 17, 2025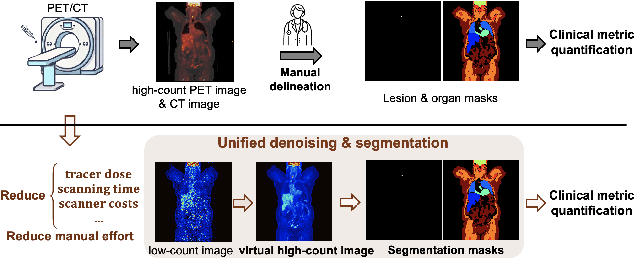
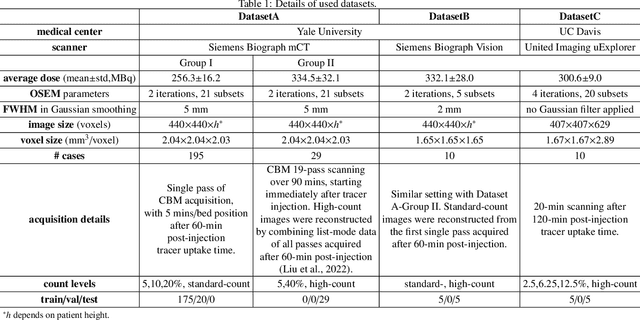
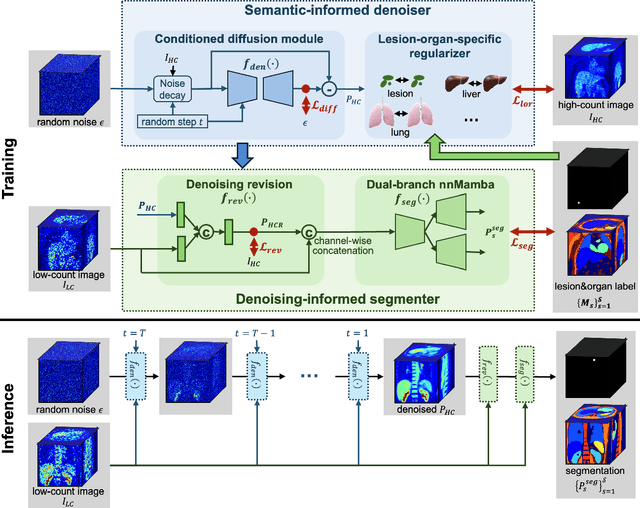
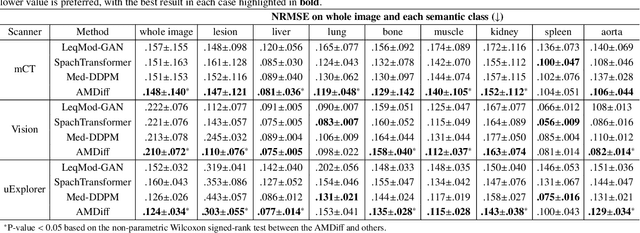
Abstract:Positron emission tomography (PET) image denoising, along with lesion and organ segmentation, are critical steps in PET-aided diagnosis. However, existing methods typically treat these tasks independently, overlooking inherent synergies between them as correlated steps in the analysis pipeline. In this work, we present the anatomically and metabolically informed diffusion (AMDiff) model, a unified framework for denoising and lesion/organ segmentation in low-count PET imaging. By integrating multi-task functionality and exploiting the mutual benefits of these tasks, AMDiff enables direct quantification of clinical metrics, such as total lesion glycolysis (TLG), from low-count inputs. The AMDiff model incorporates a semantic-informed denoiser based on diffusion strategy and a denoising-informed segmenter utilizing nnMamba architecture. The segmenter constrains denoised outputs via a lesion-organ-specific regularizer, while the denoiser enhances the segmenter by providing enriched image information through a denoising revision module. These components are connected via a warming-up mechanism to optimize multitask interactions. Experiments on multi-vendor, multi-center, and multi-noise-level datasets demonstrate the superior performance of AMDiff. For test cases below 20% of the clinical count levels from participating sites, AMDiff achieves TLG quantification biases of -26.98%, outperforming its ablated versions which yield biases of -35.85% (without the lesion-organ-specific regularizer) and -40.79% (without the denoising revision module).
Neural KEM: A Kernel Method with Deep Coefficient Prior for PET Image Reconstruction
Jan 05, 2022



Abstract:Image reconstruction of low-count positron emission tomography (PET) data is challenging. Kernel methods address the challenge by incorporating image prior information in the forward model of iterative PET image reconstruction. The kernelized expectation-maximization (KEM) algorithm has been developed and demonstrated to be effective and easy to implement. A common approach for a further improvement of the kernel method would be adding an explicit regularization, which however leads to a complex optimization problem. In this paper, we propose an implicit regularization for the kernel method by using a deep coefficient prior, which represents the kernel coefficient image in the PET forward model using a convolutional neural-network. To solve the maximum-likelihood neural network-based reconstruction problem, we apply the principle of optimization transfer to derive a neural KEM algorithm. Each iteration of the algorithm consists of two separate steps: a KEM step for image update from the projection data and a deep-learning step in the image domain for updating the kernel coefficient image using the neural network. This optimization algorithm is guaranteed to monotonically increase the data likelihood. The results from computer simulations and real patient data have demonstrated that the neural KEM can outperform existing KEM and deep image prior methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge