Rafi Brada
INFORM-CT: INtegrating LLMs and VLMs FOR Incidental Findings Management in Abdominal CT
Dec 10, 2025


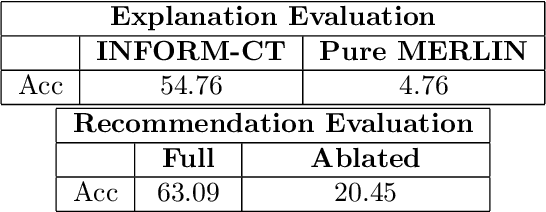
Abstract:Incidental findings in CT scans, though often benign, can have significant clinical implications and should be reported following established guidelines. Traditional manual inspection by radiologists is time-consuming and variable. This paper proposes a novel framework that leverages large language models (LLMs) and foundational vision-language models (VLMs) in a plan-and-execute agentic approach to improve the efficiency and precision of incidental findings detection, classification, and reporting for abdominal CT scans. Given medical guidelines for abdominal organs, the process of managing incidental findings is automated through a planner-executor framework. The planner, based on LLM, generates Python scripts using predefined base functions, while the executor runs these scripts to perform the necessary checks and detections, via VLMs, segmentation models, and image processing subroutines. We demonstrate the effectiveness of our approach through experiments on a CT abdominal benchmark for three organs, in a fully automatic end-to-end manner. Our results show that the proposed framework outperforms existing pure VLM-based approaches in terms of accuracy and efficiency.
Deep learning-based reconstruction of highly accelerated 3D MRI
Mar 09, 2022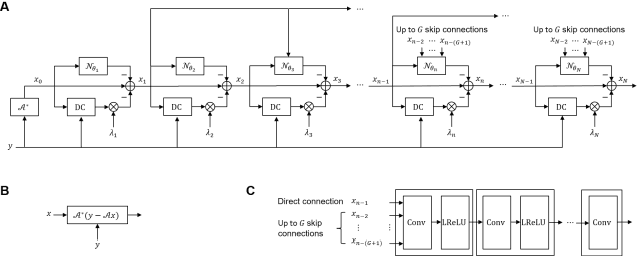
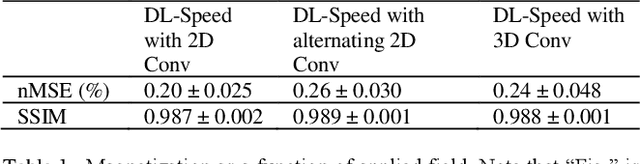

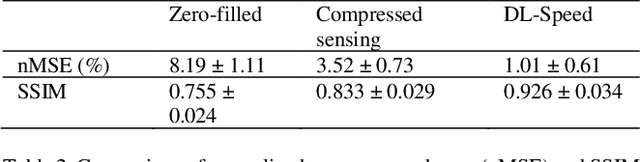
Abstract:Purpose: To accelerate brain 3D MRI scans by using a deep learning method for reconstructing images from highly-undersampled multi-coil k-space data Methods: DL-Speed, an unrolled optimization architecture with dense skip-layer connections, was trained on 3D T1-weighted brain scan data to reconstruct complex-valued images from highly-undersampled k-space data. The trained model was evaluated on 3D MPRAGE brain scan data retrospectively-undersampled with a 10-fold acceleration, compared to a conventional parallel imaging method with a 2-fold acceleration. Scores of SNR, artifacts, gray/white matter contrast, resolution/sharpness, deep gray-matter, cerebellar vermis, anterior commissure, and overall quality, on a 5-point Likert scale, were assessed by experienced radiologists. In addition, the trained model was tested on retrospectively-undersampled 3D T1-weighted LAVA (Liver Acquisition with Volume Acceleration) abdominal scan data, and prospectively-undersampled 3D MPRAGE and LAVA scans in three healthy volunteers and one, respectively. Results: The qualitative scores for DL-Speed with a 10-fold acceleration were higher than or equal to those for the parallel imaging with 2-fold acceleration. DL-Speed outperformed a compressed sensing method in quantitative metrics on retrospectively-undersampled LAVA data. DL-Speed was demonstrated to perform reasonably well on prospectively-undersampled scan data, realizing a 2-5 times reduction in scan time. Conclusion: DL-Speed was shown to accelerate 3D MPRAGE and LAVA with up to a net 10-fold acceleration, achieving 2-5 times faster scans compared to conventional parallel imaging and acceleration, while maintaining diagnostic image quality and real-time reconstruction. The brain scan data-trained DL-Speed also performed well when reconstructing abdominal LAVA scan data, demonstrating versatility of the network.
High Fidelity Deep Learning-based MRI Reconstruction with Instance-wise Discriminative Feature Matching Loss
Aug 27, 2021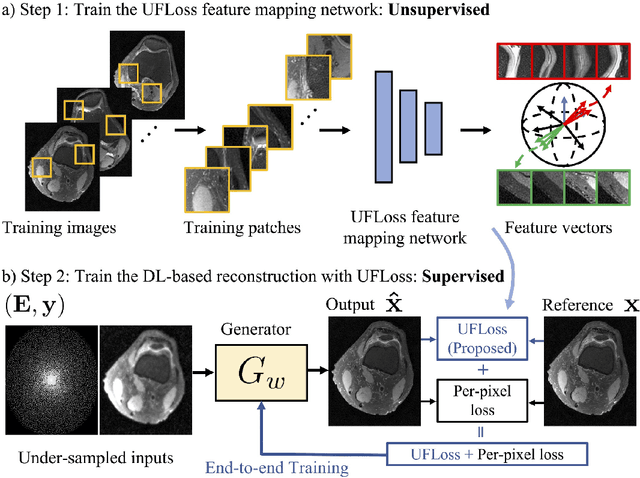
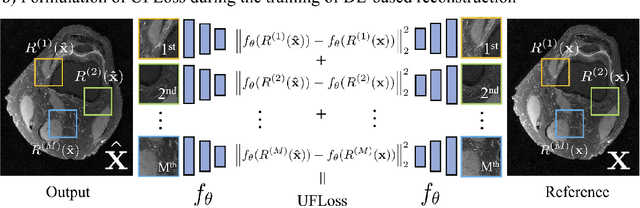
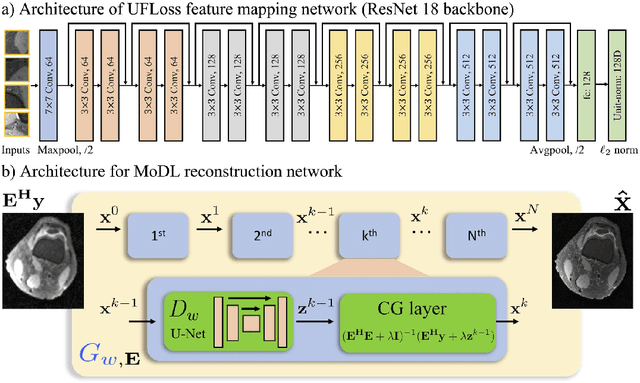
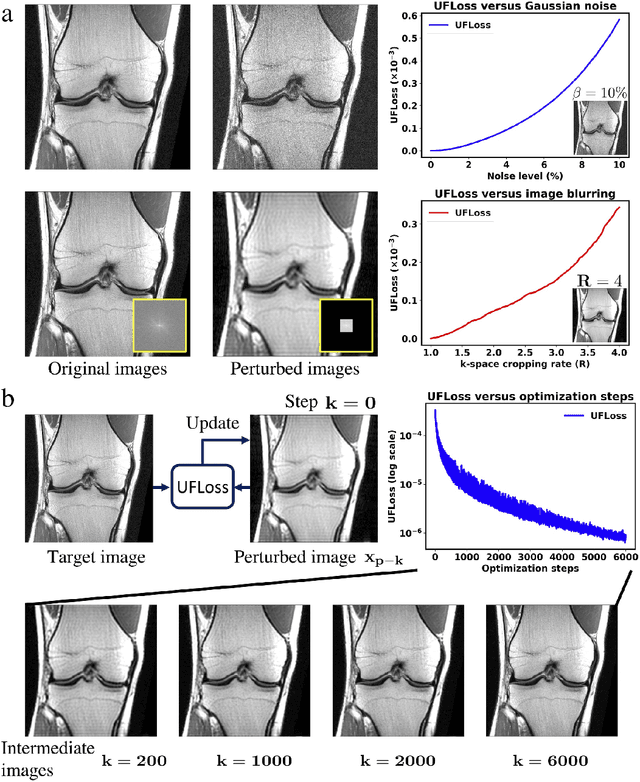
Abstract:Purpose: To improve reconstruction fidelity of fine structures and textures in deep learning (DL) based reconstructions. Methods: A novel patch-based Unsupervised Feature Loss (UFLoss) is proposed and incorporated into the training of DL-based reconstruction frameworks in order to preserve perceptual similarity and high-order statistics. The UFLoss provides instance-level discrimination by mapping similar instances to similar low-dimensional feature vectors and is trained without any human annotation. By adding an additional loss function on the low-dimensional feature space during training, the reconstruction frameworks from under-sampled or corrupted data can reproduce more realistic images that are closer to the original with finer textures, sharper edges, and improved overall image quality. The performance of the proposed UFLoss is demonstrated on unrolled networks for accelerated 2D and 3D knee MRI reconstruction with retrospective under-sampling. Quantitative metrics including NRMSE, SSIM, and our proposed UFLoss were used to evaluate the performance of the proposed method and compare it with others. Results: In-vivo experiments indicate that adding the UFLoss encourages sharper edges and more faithful contrasts compared to traditional and learning-based methods with pure l2 loss. More detailed textures can be seen in both 2D and 3D knee MR images. Quantitative results indicate that reconstruction with UFLoss can provide comparable NRMSE and a higher SSIM while achieving a much lower UFLoss value. Conclusion: We present UFLoss, a patch-based unsupervised learned feature loss, which allows the training of DL-based reconstruction to obtain more detailed texture, finer features, and sharper edges with higher overall image quality under DL-based reconstruction frameworks.
A Novel Approach for Correcting Multiple Discrete Rigid In-Plane Motions Artefacts in MRI Scans
Jun 29, 2020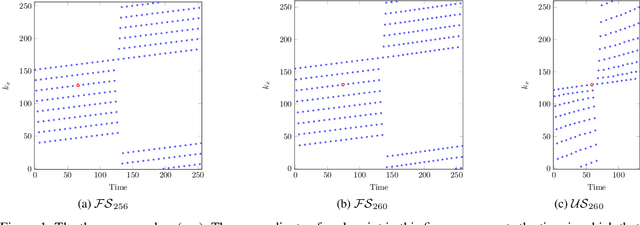
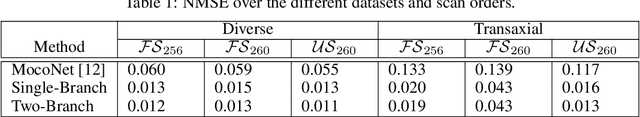
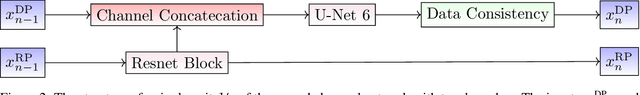
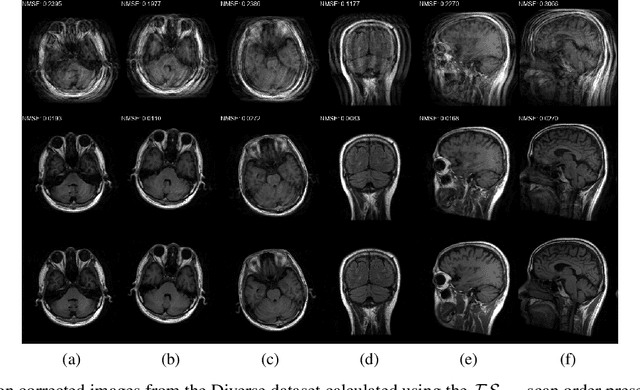
Abstract:Motion artefacts created by patient motion during an MRI scan occur frequently in practice, often rendering the scans clinically unusable and requiring a re-scan. While many methods have been employed to ameliorate the effects of patient motion, these often fall short in practice. In this paper we propose a novel method for removing motion artefacts using a deep neural network with two input branches that discriminates between patient poses using the motion's timing. The first branch receives a subset of the $k$-space data collected during a single patient pose, and the second branch receives the remaining part of the collected $k$-space data. The proposed method can be applied to artefacts generated by multiple movements of the patient. Furthermore, it can be used to correct motion for the case where $k$-space has been under-sampled, to shorten the scan time, as is common when using methods such as parallel imaging or compressed sensing. Experimental results on both simulated and real MRI data show the efficacy of our approach.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge