Rafael Izbicki
CP4SBI: Local Conformal Calibration of Credible Sets in Simulation-Based Inference
Aug 23, 2025Abstract:Current experimental scientists have been increasingly relying on simulation-based inference (SBI) to invert complex non-linear models with intractable likelihoods. However, posterior approximations obtained with SBI are often miscalibrated, causing credible regions to undercover true parameters. We develop $\texttt{CP4SBI}$, a model-agnostic conformal calibration framework that constructs credible sets with local Bayesian coverage. Our two proposed variants, namely local calibration via regression trees and CDF-based calibration, enable finite-sample local coverage guarantees for any scoring function, including HPD, symmetric, and quantile-based regions. Experiments on widely used SBI benchmarks demonstrate that our approach improves the quality of uncertainty quantification for neural posterior estimators using both normalizing flows and score-diffusion modeling.
Epistemic Uncertainty in Conformal Scores: A Unified Approach
Feb 10, 2025Abstract:Conformal prediction methods create prediction bands with distribution-free guarantees but do not explicitly capture epistemic uncertainty, which can lead to overconfident predictions in data-sparse regions. Although recent conformal scores have been developed to address this limitation, they are typically designed for specific tasks, such as regression or quantile regression. Moreover, they rely on particular modeling choices for epistemic uncertainty, restricting their applicability. We introduce $\texttt{EPICSCORE}$, a model-agnostic approach that enhances any conformal score by explicitly integrating epistemic uncertainty. Leveraging Bayesian techniques such as Gaussian Processes, Monte Carlo Dropout, or Bayesian Additive Regression Trees, $\texttt{EPICSCORE}$ adaptively expands predictive intervals in regions with limited data while maintaining compact intervals where data is abundant. As with any conformal method, it preserves finite-sample marginal coverage. Additionally, it also achieves asymptotic conditional coverage. Experiments demonstrate its good performance compared to existing methods. Designed for compatibility with any Bayesian model, but equipped with distribution-free guarantees, $\texttt{EPICSCORE}$ provides a general-purpose framework for uncertainty quantification in prediction problems.
Distribution-Free Calibration of Statistical Confidence Sets
Nov 28, 2024



Abstract:Constructing valid confidence sets is a crucial task in statistical inference, yet traditional methods often face challenges when dealing with complex models or limited observed sample sizes. These challenges are frequently encountered in modern applications, such as Likelihood-Free Inference (LFI). In these settings, confidence sets may fail to maintain a confidence level close to the nominal value. In this paper, we introduce two novel methods, TRUST and TRUST++, for calibrating confidence sets to achieve distribution-free conditional coverage. These methods rely entirely on simulated data from the statistical model to perform calibration. Leveraging insights from conformal prediction techniques adapted to the statistical inference context, our methods ensure both finite-sample local coverage and asymptotic conditional coverage as the number of simulations increases, even if n is small. They effectively handle nuisance parameters and provide computationally efficient uncertainty quantification for the estimated confidence sets. This allows users to assess whether additional simulations are necessary for robust inference. Through theoretical analysis and experiments on models with both tractable and intractable likelihoods, we demonstrate that our methods outperform existing approaches, particularly in small-sample regimes. This work bridges the gap between conformal prediction and statistical inference, offering practical tools for constructing valid confidence sets in complex models.
PersonalizedUS: Interpretable Breast Cancer Risk Assessment with Local Coverage Uncertainty Quantification
Aug 28, 2024



Abstract:Correctly assessing the malignancy of breast lesions identified during ultrasound examinations is crucial for effective clinical decision-making. However, the current "golden standard" relies on manual BI-RADS scoring by clinicians, often leading to unnecessary biopsies and a significant mental health burden on patients and their families. In this paper, we introduce PersonalizedUS, an interpretable machine learning system that leverages recent advances in conformal prediction to provide precise and personalized risk estimates with local coverage guarantees and sensitivity, specificity, and predictive values above 0.9 across various threshold levels. In particular, we identify meaningful lesion subgroups where distribution-free, model-agnostic conditional coverage holds, with approximately 90% of our prediction sets containing only the ground truth in most lesion subgroups, thus explicitly characterizing for which patients the model is most suitably applied. Moreover, we make available a curated tabular dataset of 1936 biopsied breast lesions from a recent observational multicenter study and benchmark the performance of several state-of-the-art learning algorithms. We also report a successful case study of the deployed system in the same multicenter context. Concrete clinical benefits include up to a 65% reduction in requested biopsies among BI-RADS 4a and 4b lesions, with minimal to no missed cancer cases.
Regression Trees for Fast and Adaptive Prediction Intervals
Feb 13, 2024
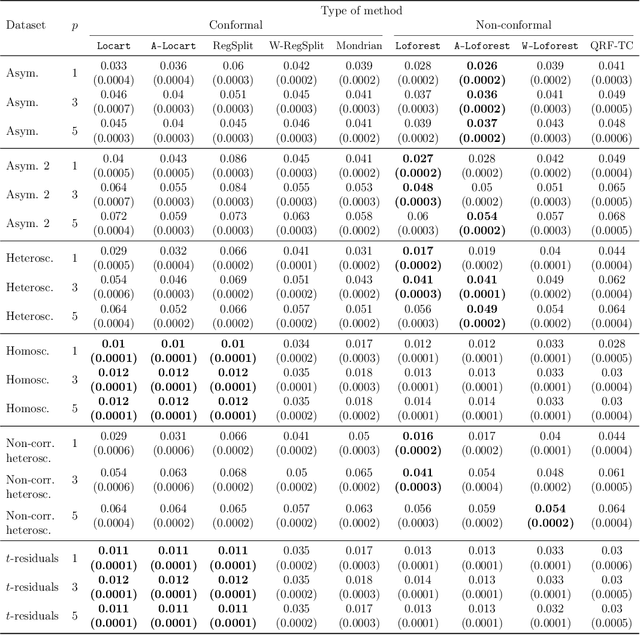


Abstract:Predictive models make mistakes. Hence, there is a need to quantify the uncertainty associated with their predictions. Conformal inference has emerged as a powerful tool to create statistically valid prediction regions around point predictions, but its naive application to regression problems yields non-adaptive regions. New conformal scores, often relying upon quantile regressors or conditional density estimators, aim to address this limitation. Although they are useful for creating prediction bands, these scores are detached from the original goal of quantifying the uncertainty around an arbitrary predictive model. This paper presents a new, model-agnostic family of methods to calibrate prediction intervals for regression problems with local coverage guarantees. Our approach is based on pursuing the coarsest partition of the feature space that approximates conditional coverage. We create this partition by training regression trees and Random Forests on conformity scores. Our proposal is versatile, as it applies to various conformity scores and prediction settings and demonstrates superior scalability and performance compared to established baselines in simulated and real-world datasets. We provide a Python package clover that implements our methods using the standard scikit-learn interface.
Classification under Nuisance Parameters and Generalized Label Shift in Likelihood-Free Inference
Feb 08, 2024



Abstract:An open scientific challenge is how to classify events with reliable measures of uncertainty, when we have a mechanistic model of the data-generating process but the distribution over both labels and latent nuisance parameters is different between train and target data. We refer to this type of distributional shift as generalized label shift (GLS). Direct classification using observed data $\mathbf{X}$ as covariates leads to biased predictions and invalid uncertainty estimates of labels $Y$. We overcome these biases by proposing a new method for robust uncertainty quantification that casts classification as a hypothesis testing problem under nuisance parameters. The key idea is to estimate the classifier's receiver operating characteristic (ROC) across the entire nuisance parameter space, which allows us to devise cutoffs that are invariant under GLS. Our method effectively endows a pre-trained classifier with domain adaptation capabilities and returns valid prediction sets while maintaining high power. We demonstrate its performance on two challenging scientific problems in biology and astroparticle physics with data from realistic mechanistic models.
Distribution-Free Conformal Joint Prediction Regions for Neural Marked Temporal Point Processes
Jan 09, 2024



Abstract:Sequences of labeled events observed at irregular intervals in continuous time are ubiquitous across various fields. Temporal Point Processes (TPPs) provide a mathematical framework for modeling these sequences, enabling inferences such as predicting the arrival time of future events and their associated label, called mark. However, due to model misspecification or lack of training data, these probabilistic models may provide a poor approximation of the true, unknown underlying process, with prediction regions extracted from them being unreliable estimates of the underlying uncertainty. This paper develops more reliable methods for uncertainty quantification in neural TPP models via the framework of conformal prediction. A primary objective is to generate a distribution-free joint prediction region for the arrival time and mark, with a finite-sample marginal coverage guarantee. A key challenge is to handle both a strictly positive, continuous response and a categorical response, without distributional assumptions. We first consider a simple but overly conservative approach that combines individual prediction regions for the event arrival time and mark. Then, we introduce a more effective method based on bivariate highest density regions derived from the joint predictive density of event arrival time and mark. By leveraging the dependencies between these two variables, this method exclude unlikely combinations of the two, resulting in sharper prediction regions while still attaining the pre-specified coverage level. We also explore the generation of individual univariate prediction regions for arrival times and marks through conformal regression and classification techniques. Moreover, we investigate the stronger notion of conditional coverage. Finally, through extensive experimentation on both simulated and real-world datasets, we assess the validity and efficiency of these methods.
Expertise-based Weighting for Regression Models with Noisy Labels
May 12, 2023Abstract:Regression methods assume that accurate labels are available for training. However, in certain scenarios, obtaining accurate labels may not be feasible, and relying on multiple specialists with differing opinions becomes necessary. Existing approaches addressing noisy labels often impose restrictive assumptions on the regression function. In contrast, this paper presents a novel, more flexible approach. Our method consists of two steps: estimating each labeler's expertise and combining their opinions using learned weights. We then regress the weighted average against the input features to build the prediction model. The proposed method is formally justified and empirically demonstrated to outperform existing techniques on simulated and real data. Furthermore, its flexibility enables the utilization of any machine learning technique in both steps. In summary, this method offers a simple, fast, and effective solution for training regression models with noisy labels derived from diverse expert opinions.
Is augmentation effective to improve prediction in imbalanced text datasets?
Apr 20, 2023



Abstract:Imbalanced datasets present a significant challenge for machine learning models, often leading to biased predictions. To address this issue, data augmentation techniques are widely used in natural language processing (NLP) to generate new samples for the minority class. However, in this paper, we challenge the common assumption that data augmentation is always necessary to improve predictions on imbalanced datasets. Instead, we argue that adjusting the classifier cutoffs without data augmentation can produce similar results to oversampling techniques. Our study provides theoretical and empirical evidence to support this claim. Our findings contribute to a better understanding of the strengths and limitations of different approaches to dealing with imbalanced data, and help researchers and practitioners make informed decisions about which methods to use for a given task.
Flexible conditional density estimation for time series
Jan 23, 2023Abstract:This paper introduces FlexCodeTS, a new conditional density estimator for time series. FlexCodeTS is a flexible nonparametric conditional density estimator, which can be based on an arbitrary regression method. It is shown that FlexCodeTS inherits the rate of convergence of the chosen regression method. Hence, FlexCodeTS can adapt its convergence by employing the regression method that best fits the structure of data. From an empirical perspective, FlexCodeTS is compared to NNKCDE and GARCH in both simulated and real data. FlexCodeTS is shown to generally obtain the best performance among the selected methods according to either the CDE loss or the pinball loss.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge