Raúl San José Estépar
$\texttt{NePhi}$: Neural Deformation Fields for Approximately Diffeomorphic Medical Image Registration
Sep 13, 2023
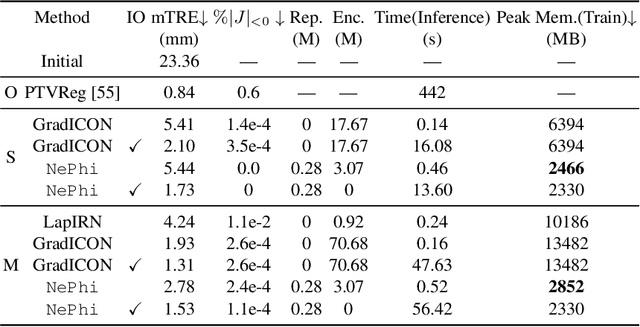

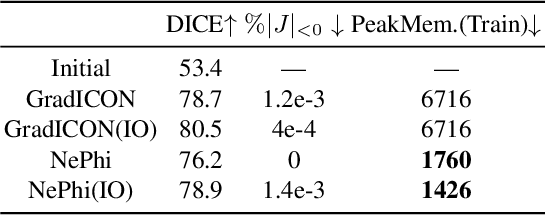
Abstract:This work proposes $\texttt{NePhi}$, a neural deformation model which results in approximately diffeomorphic transformations. In contrast to the predominant voxel-based approaches, $\texttt{NePhi}$ represents deformations functionally which allows for memory-efficient training and inference. This is of particular importance for large volumetric registrations. Further, while medical image registration approaches representing transformation maps via multi-layer perceptrons have been proposed, $\texttt{NePhi}$ facilitates both pairwise optimization-based registration $\textit{as well as}$ learning-based registration via predicted or optimized global and local latent codes. Lastly, as deformation regularity is a highly desirable property for most medical image registration tasks, $\texttt{NePhi}$ makes use of gradient inverse consistency regularization which empirically results in approximately diffeomorphic transformations. We show the performance of $\texttt{NePhi}$ on two 2D synthetic datasets as well as on real 3D lung registration. Our results show that $\texttt{NePhi}$ can achieve similar accuracies as voxel-based representations in a single-resolution registration setting while using less memory and allowing for faster instance-optimization.
GradICON: Approximate Diffeomorphisms via Gradient Inverse Consistency
Jun 13, 2022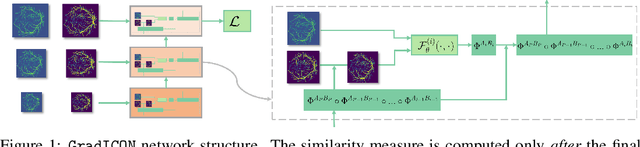
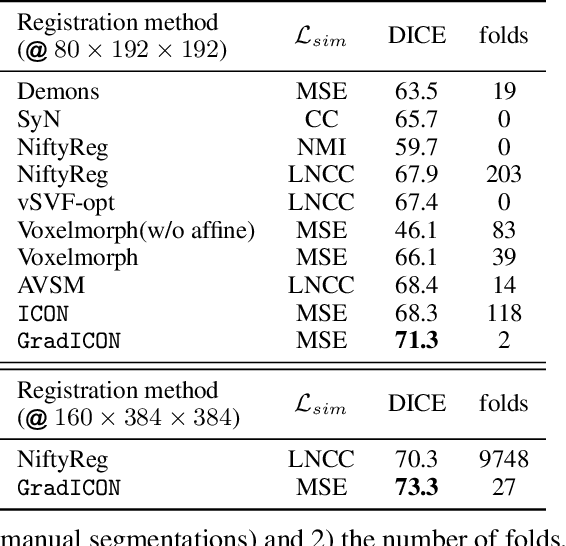
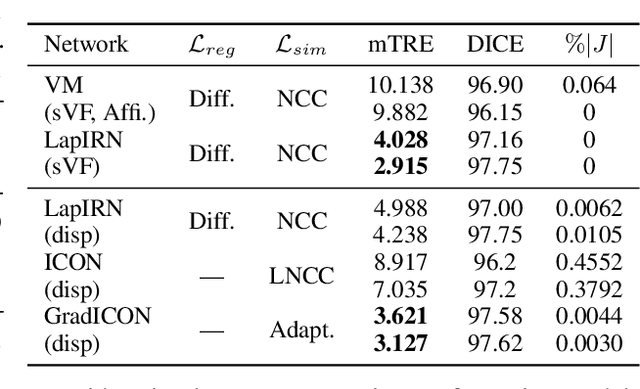
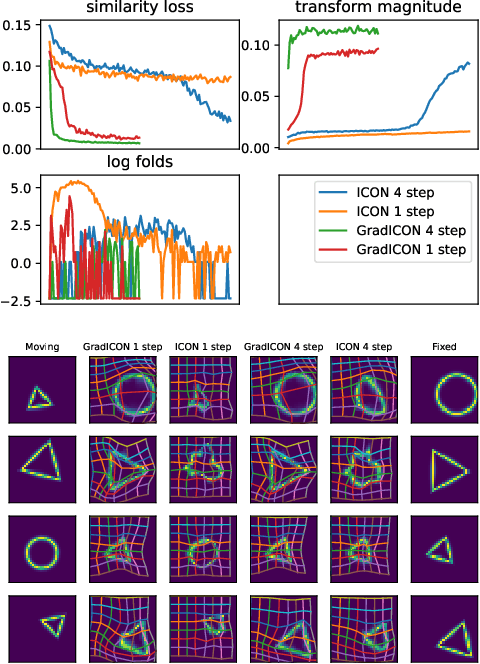
Abstract:Many registration approaches exist with early work focusing on optimization-based approaches for image pairs. Recent work focuses on deep registration networks to predict spatial transformations. In both cases, commonly used non-parametric registration models, which estimate transformation functions instead of low-dimensional transformation parameters, require choosing a suitable regularizer (to encourage smooth transformations) and its parameters. This makes models difficult to tune and restricts deformations to the deformation space permissible by the chosen regularizer. While deep-learning models for optical flow exist that do not regularize transformations and instead entirely rely on the data these might not yield diffeomorphic transformations which are desirable for medical image registration. In this work, we therefore develop GradICON building upon the unsupervised ICON deep-learning registration approach, which only uses inverse-consistency for regularization. However, in contrast to ICON, we prove and empirically verify that using a gradient inverse-consistency loss not only significantly improves convergence, but also results in a similar implicit regularization of the resulting transformation map. Synthetic experiments and experiments on magnetic resonance (MR) knee images and computed tomography (CT) lung images show the excellent performance of GradICON. We achieve state-of-the-art (SOTA) accuracy while retaining a simple registration formulation, which is practically important.
LiftReg: Limited Angle 2D/3D Deformable Registration
Mar 10, 2022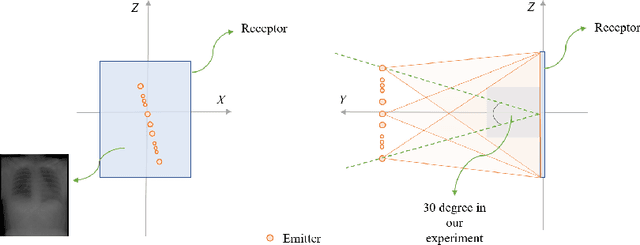
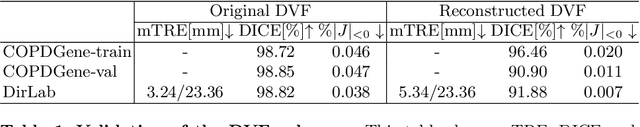
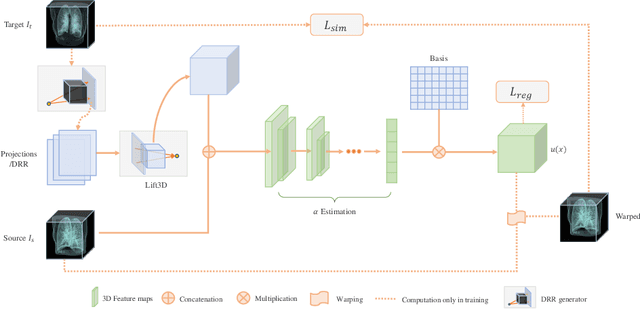
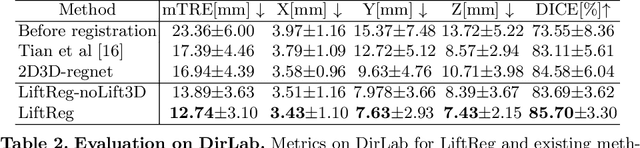
Abstract:We propose LiftReg, a 2D/3D deformable registration approach. LiftReg is a deep registration framework which is trained using sets of digitally reconstructed radiographs (DRR) and computed tomography (CT) image pairs. By using simulated training data, LiftReg can use a high-quality CT-CT image similarity measure, which helps the network to learn a high-quality deformation space. To further improve registration quality and to address the inherent depth ambiguities of very limited angle acquisitions, we propose to use features extracted from the backprojected 2D images and a statistical deformation model. We test our approach on the DirLab lung registration dataset and show that it outperforms an existing learning-based pairwise registration approach.
Computer Aided Detection for Pulmonary Embolism Challenge (CAD-PE)
Mar 30, 2020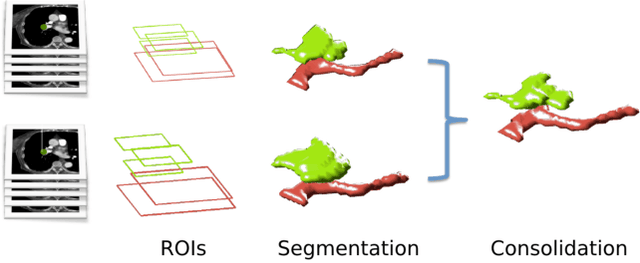
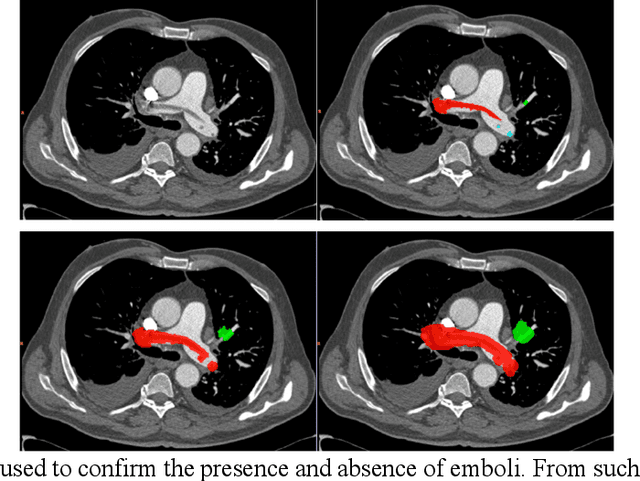
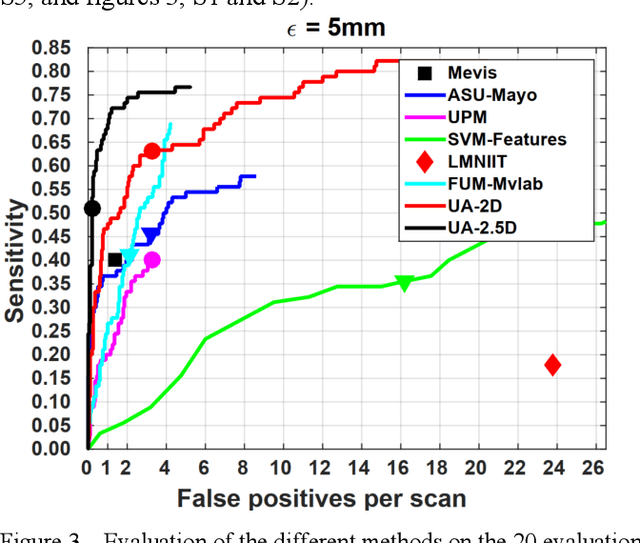
Abstract:Rationale: Computer aided detection (CAD) algorithms for Pulmonary Embolism (PE) algorithms have been shown to increase radiologists' sensitivity with a small increase in specificity. However, CAD for PE has not been adopted into clinical practice, likely because of the high number of false positives current CAD software produces. Objective: To generate a database of annotated computed tomography pulmonary angiographies, use it to compare the sensitivity and false positive rate of current algorithms and to develop new methods that improve such metrics. Methods: 91 Computed tomography pulmonary angiography scans were annotated by at least one radiologist by segmenting all pulmonary emboli visible on the study. 20 annotated CTPAs were open to the public in the form of a medical image analysis challenge. 20 more were kept for evaluation purposes. 51 were made available post-challenge. 8 submissions, 6 of them novel, were evaluated on the 20 evaluation CTPAs. Performance was measured as per embolus sensitivity vs. false positives per scan curve. Results: The best algorithms achieved a per-embolus sensitivity of 75% at 2 false positives per scan (fps) or of 70% at 1 fps, outperforming the state of the art. Deep learning approaches outperformed traditional machine learning ones, and their performance improved with the number of training cases. Significance: Through this work and challenge we have improved the state-of-the art of computer aided detection algorithms for pulmonary embolism. An open database and an evaluation benchmark for such algorithms have been generated, easing the development of further improvements. Implications on clinical practice will need further research.
Generative-based Airway and Vessel Morphology Quantification on Chest CT Images
Mar 13, 2020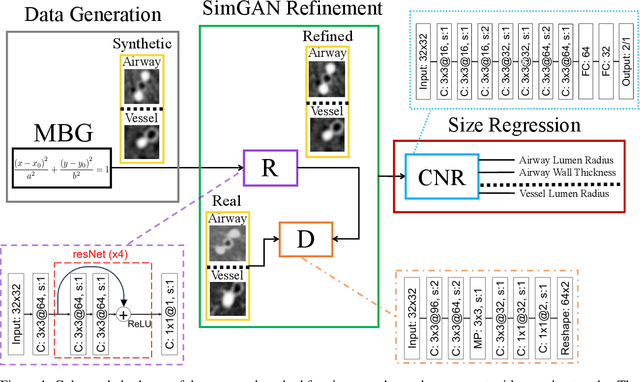
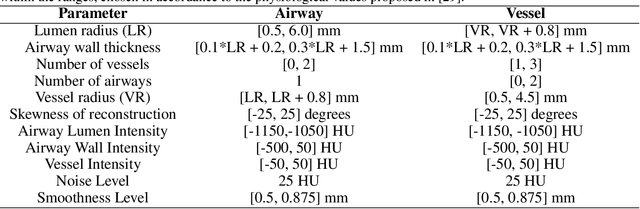
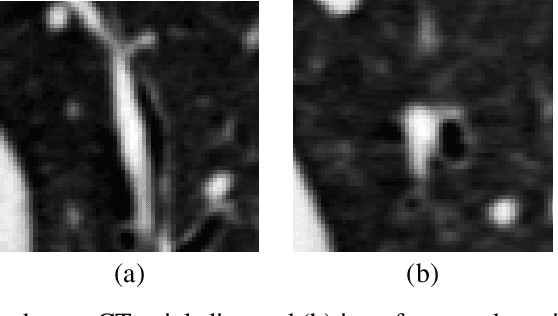
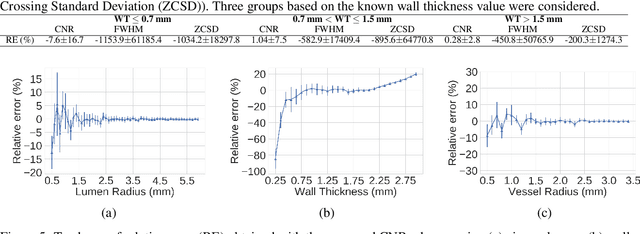
Abstract:Accurately and precisely characterizing the morphology of small pulmonary structures from Computed Tomography (CT) images, such as airways and vessels, is becoming of great importance for diagnosis of pulmonary diseases. The smaller conducting airways are the major site of increased airflow resistance in chronic obstructive pulmonary disease (COPD), while accurately sizing vessels can help identify arterial and venous changes in lung regions that may determine future disorders. However, traditional methods are often limited due to image resolution and artifacts. We propose a Convolutional Neural Regressor (CNR) that provides cross-sectional measurement of airway lumen, airway wall thickness, and vessel radius. CNR is trained with data created by a generative model of synthetic structures which is used in combination with Simulated and Unsupervised Generative Adversarial Network (SimGAN) to create simulated and refined airways and vessels with known ground-truth. For validation, we first use synthetically generated airways and vessels produced by the proposed generative model to compute the relative error and directly evaluate the accuracy of CNR in comparison with traditional methods. Then, in-vivo validation is performed by analyzing the association between the percentage of the predicted forced expiratory volume in one second (FEV1\%) and the value of the Pi10 parameter, two well-known measures of lung function and airway disease, for airways. For vessels, we assess the correlation between our estimate of the small-vessel blood volume and the lungs' diffusing capacity for carbon monoxide (DLCO). The results demonstrate that Convolutional Neural Networks (CNNs) provide a promising direction for accurately measuring vessels and airways on chest CT images with physiological correlates.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge