Germán González
UMLS-ChestNet: A deep convolutional neural network for radiological findings, differential diagnoses and localizations of COVID-19 in chest x-rays
Jun 06, 2020



Abstract:In this work we present a method for the detection of radiological findings, their location and differential diagnoses from chest x-rays. Unlike prior works that focus on the detection of few pathologies, we use a hierarchical taxonomy mapped to the Unified Medical Language System (UMLS) terminology to identify 189 radiological findings, 22 differential diagnosis and 122 anatomic locations, including ground glass opacities, infiltrates, consolidations and other radiological findings compatible with COVID-19. We train the system on one large database of 92,594 frontal chest x-rays (AP or PA, standing, supine or decubitus) and a second database of 2,065 frontal images of COVID-19 patients identified by at least one positive Polymerase Chain Reaction (PCR) test. The reference labels are obtained through natural language processing of the radiological reports. On 23,159 test images, the proposed neural network obtains an AUC of 0.94 for the diagnosis of COVID-19. To our knowledge, this work uses the largest chest x-ray dataset of COVID-19 positive cases to date and is the first one to use a hierarchical labeling schema and to provide interpretability of the results, not only by using network attention methods, but also by indicating the radiological findings that have led to the diagnosis.
BIMCV COVID-19+: a large annotated dataset of RX and CT images from COVID-19 patients
Jun 05, 2020



Abstract:This paper describes BIMCV COVID-19+, a large dataset from the Valencian Region Medical ImageBank (BIMCV) containing chest X-ray images CXR (CR, DX) and computed tomography (CT) imaging of COVID-19+ patients along with their radiological findings and locations, pathologies, radiological reports (in Spanish), DICOM metadata, Polymerase chain reaction (PCR), Immunoglobulin G (IgG) and Immunoglobulin M (IgM) diagnostic antibody tests. The findings have been mapped onto standard Unified Medical Language System (UMLS) terminology and cover a wide spectrum of thoracic entities, unlike the considerably more reduced number of entities annotated in previous datasets. Images are stored in high resolution and entities are localized with anatomical labels and stored in a Medical Imaging Data Structure (MIDS) format. In addition, 10 images were annotated by a team of radiologists to include semantic segmentation of radiological findings. This first iteration of the database includes 1,380 CX, 885 DX and 163 CT studies from 1,311 COVID-19+ patients. This is, to the best of our knowledge, the largest COVID-19+ dataset of images available in an open format. The dataset can be downloaded from http://bimcv.cipf.es/bimcv-projects/bimcv-covid19.
Computer Aided Detection for Pulmonary Embolism Challenge (CAD-PE)
Mar 30, 2020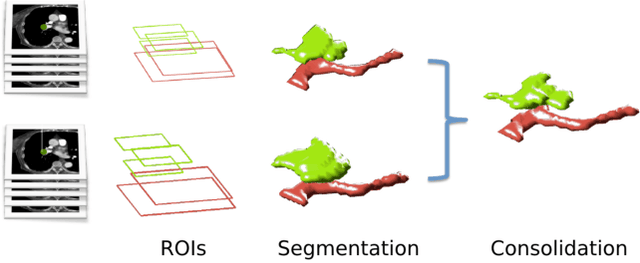
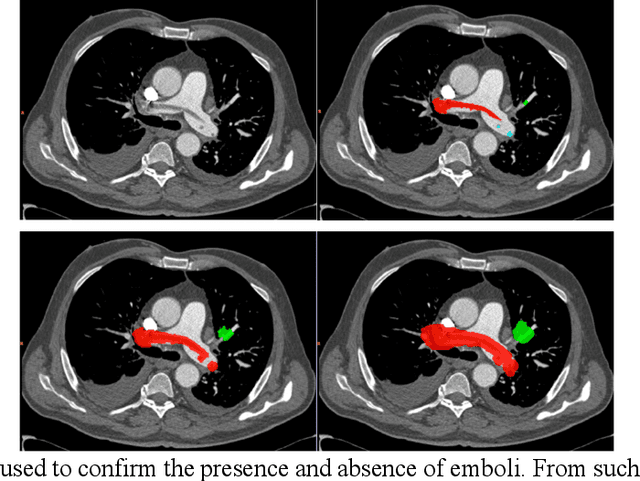
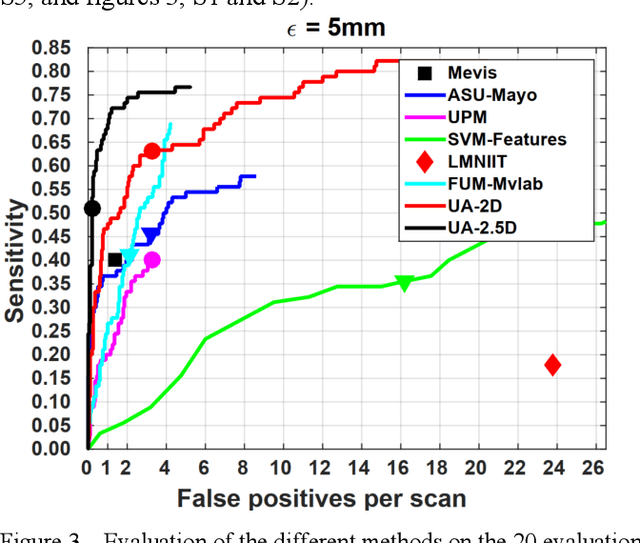
Abstract:Rationale: Computer aided detection (CAD) algorithms for Pulmonary Embolism (PE) algorithms have been shown to increase radiologists' sensitivity with a small increase in specificity. However, CAD for PE has not been adopted into clinical practice, likely because of the high number of false positives current CAD software produces. Objective: To generate a database of annotated computed tomography pulmonary angiographies, use it to compare the sensitivity and false positive rate of current algorithms and to develop new methods that improve such metrics. Methods: 91 Computed tomography pulmonary angiography scans were annotated by at least one radiologist by segmenting all pulmonary emboli visible on the study. 20 annotated CTPAs were open to the public in the form of a medical image analysis challenge. 20 more were kept for evaluation purposes. 51 were made available post-challenge. 8 submissions, 6 of them novel, were evaluated on the 20 evaluation CTPAs. Performance was measured as per embolus sensitivity vs. false positives per scan curve. Results: The best algorithms achieved a per-embolus sensitivity of 75% at 2 false positives per scan (fps) or of 70% at 1 fps, outperforming the state of the art. Deep learning approaches outperformed traditional machine learning ones, and their performance improved with the number of training cases. Significance: Through this work and challenge we have improved the state-of-the art of computer aided detection algorithms for pulmonary embolism. An open database and an evaluation benchmark for such algorithms have been generated, easing the development of further improvements. Implications on clinical practice will need further research.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge