Antonio Pertusa
PadChest-GR: A Bilingual Chest X-ray Dataset for Grounded Radiology Report Generation
Nov 07, 2024



Abstract:Radiology report generation (RRG) aims to create free-text radiology reports from clinical imaging. Grounded radiology report generation (GRRG) extends RRG by including the localisation of individual findings on the image. Currently, there are no manually annotated chest X-ray (CXR) datasets to train GRRG models. In this work, we present a dataset called PadChest-GR (Grounded-Reporting) derived from PadChest aimed at training GRRG models for CXR images. We curate a public bi-lingual dataset of 4,555 CXR studies with grounded reports (3,099 abnormal and 1,456 normal), each containing complete lists of sentences describing individual present (positive) and absent (negative) findings in English and Spanish. In total, PadChest-GR contains 7,037 positive and 3,422 negative finding sentences. Every positive finding sentence is associated with up to two independent sets of bounding boxes labelled by different readers and has categorical labels for finding type, locations, and progression. To the best of our knowledge, PadChest-GR is the first manually curated dataset designed to train GRRG models for understanding and interpreting radiological images and generated text. By including detailed localization and comprehensive annotations of all clinically relevant findings, it provides a valuable resource for developing and evaluating GRRG models from CXR images. PadChest-GR can be downloaded under request from https://bimcv.cipf.es/bimcv-projects/padchest-gr/
Few-shot learning for COVID-19 Chest X-Ray Classification with Imbalanced Data: An Inter vs. Intra Domain Study
Jan 18, 2024Abstract:Medical image datasets are essential for training models used in computer-aided diagnosis, treatment planning, and medical research. However, some challenges are associated with these datasets, including variability in data distribution, data scarcity, and transfer learning issues when using models pre-trained from generic images. This work studies the effect of these challenges at the intra- and inter-domain level in few-shot learning scenarios with severe data imbalance. For this, we propose a methodology based on Siamese neural networks in which a series of techniques are integrated to mitigate the effects of data scarcity and distribution imbalance. Specifically, different initialization and data augmentation methods are analyzed, and four adaptations to Siamese networks of solutions to deal with imbalanced data are introduced, including data balancing and weighted loss, both separately and combined, and with a different balance of pairing ratios. Moreover, we also assess the inference process considering four classifiers, namely Histogram, $k$NN, SVM, and Random Forest. Evaluation is performed on three chest X-ray datasets with annotated cases of both positive and negative COVID-19 diagnoses. The accuracy of each technique proposed for the Siamese architecture is analyzed separately and their results are compared to those obtained using equivalent methods on a state-of-the-art CNN. We conclude that the introduced techniques offer promising improvements over the baseline in almost all cases, and that the selection of the technique may vary depending on the amount of data available and the level of imbalance.
Multi-Label Logo Recognition and Retrieval based on Weighted Fusion of Neural Features
May 11, 2022

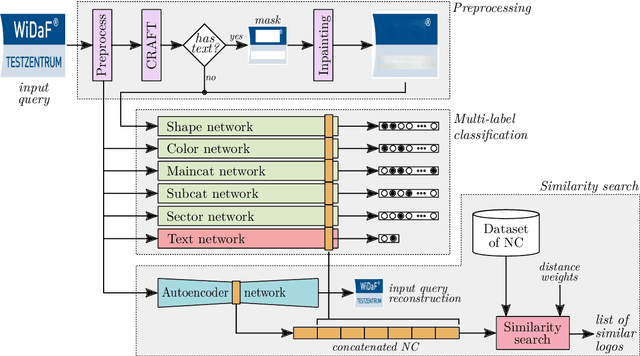

Abstract:Logo classification is a particular case of image classification, since these may contain only text, images, or a combination of both. In this work, we propose a system for the multi-label classification and similarity search of logo images. The method allows obtaining the most similar logos on the basis of their shape, color, business sector, semantics, general characteristics, or a combination of such features established by the user. This is done by employing a set of multi-label networks specialized in certain characteristics of logos. The features extracted from these networks are combined to perform the similarity search according to the search criteria established. Since the text of logos is sometimes irrelevant for the classification, a preprocessing stage is carried out to remove it, thus improving the overall performance. The proposed approach is evaluated using the European Union Trademark (EUTM) dataset, structured with the hierarchical Vienna classification system, which includes a series of metadata with which to index trademarks. We also make a comparison between well known logo topologies and Vienna in order to help designers understand their correspondences. The experimentation carried out attained reliable performance results, both quantitatively and qualitatively, which outperformed the state-of-the-art results. In addition, since the semantics and classification of brands can often be subjective, we also surveyed graphic design students and professionals in order to assess the reliability of the proposed method.
UMLS-ChestNet: A deep convolutional neural network for radiological findings, differential diagnoses and localizations of COVID-19 in chest x-rays
Jun 06, 2020



Abstract:In this work we present a method for the detection of radiological findings, their location and differential diagnoses from chest x-rays. Unlike prior works that focus on the detection of few pathologies, we use a hierarchical taxonomy mapped to the Unified Medical Language System (UMLS) terminology to identify 189 radiological findings, 22 differential diagnosis and 122 anatomic locations, including ground glass opacities, infiltrates, consolidations and other radiological findings compatible with COVID-19. We train the system on one large database of 92,594 frontal chest x-rays (AP or PA, standing, supine or decubitus) and a second database of 2,065 frontal images of COVID-19 patients identified by at least one positive Polymerase Chain Reaction (PCR) test. The reference labels are obtained through natural language processing of the radiological reports. On 23,159 test images, the proposed neural network obtains an AUC of 0.94 for the diagnosis of COVID-19. To our knowledge, this work uses the largest chest x-ray dataset of COVID-19 positive cases to date and is the first one to use a hierarchical labeling schema and to provide interpretability of the results, not only by using network attention methods, but also by indicating the radiological findings that have led to the diagnosis.
BIMCV COVID-19+: a large annotated dataset of RX and CT images from COVID-19 patients
Jun 05, 2020



Abstract:This paper describes BIMCV COVID-19+, a large dataset from the Valencian Region Medical ImageBank (BIMCV) containing chest X-ray images CXR (CR, DX) and computed tomography (CT) imaging of COVID-19+ patients along with their radiological findings and locations, pathologies, radiological reports (in Spanish), DICOM metadata, Polymerase chain reaction (PCR), Immunoglobulin G (IgG) and Immunoglobulin M (IgM) diagnostic antibody tests. The findings have been mapped onto standard Unified Medical Language System (UMLS) terminology and cover a wide spectrum of thoracic entities, unlike the considerably more reduced number of entities annotated in previous datasets. Images are stored in high resolution and entities are localized with anatomical labels and stored in a Medical Imaging Data Structure (MIDS) format. In addition, 10 images were annotated by a team of radiologists to include semantic segmentation of radiological findings. This first iteration of the database includes 1,380 CX, 885 DX and 163 CT studies from 1,311 COVID-19+ patients. This is, to the best of our knowledge, the largest COVID-19+ dataset of images available in an open format. The dataset can be downloaded from http://bimcv.cipf.es/bimcv-projects/bimcv-covid19.
A holistic approach to polyphonic music transcription with neural networks
Oct 26, 2019



Abstract:We present a framework based on neural networks to extract music scores directly from polyphonic audio in an end-to-end fashion. Most previous Automatic Music Transcription (AMT) methods seek a piano-roll representation of the pitches, that can be further transformed into a score by incorporating tempo estimation, beat tracking, key estimation or rhythm quantization. Unlike these methods, our approach generates music notation directly from the input audio in a single stage. For this, we use a Convolutional Recurrent Neural Network (CRNN) with Connectionist Temporal Classification (CTC) loss function which does not require annotated alignments of audio frames with the score rhythmic information. We trained our model using as input Haydn, Mozart, and Beethoven string quartets and Bach chorales synthesized with different tempos and expressive performances. The output is a textual representation of four-voice music scores based on **kern format. Although the proposed approach is evaluated in a simplified scenario, results show that this model can learn to transcribe scores directly from audio signals, opening a promising avenue towards complete AMT.
PadChest: A large chest x-ray image dataset with multi-label annotated reports
Feb 07, 2019



Abstract:We present a labeled large-scale, high resolution chest x-ray dataset for the automated exploration of medical images along with their associated reports. This dataset includes more than 160,000 images obtained from 67,000 patients that were interpreted and reported by radiologists at Hospital San Juan Hospital (Spain) from 2009 to 2017, covering six different position views and additional information on image acquisition and patient demography. The reports were labeled with 174 different radiographic findings, 19 differential diagnoses and 104 anatomic locations organized as a hierarchical taxonomy and mapped onto standard Unified Medical Language System (UMLS) terminology. Of these reports, 27% were manually annotated by trained physicians and the remaining set was labeled using a supervised method based on a recurrent neural network with attention mechanisms. The labels generated were then validated in an independent test set achieving a 0.93 Micro-F1 score. To the best of our knowledge, this is one of the largest public chest x-ray database suitable for training supervised models concerning radiographs, and the first to contain radiographic reports in Spanish. The PadChest dataset can be downloaded from http://bimcv.cipf.es/bimcv-projects/padchest/.
Learning Eligibility in Cancer Clinical Trials using Deep Neural Networks
Jul 25, 2018



Abstract:Interventional cancer clinical trials are generally too restrictive, and some patients are often excluded on the basis of comorbidity, past or concomitant treatments, or the fact that they are over a certain age. The efficacy and safety of new treatments for patients with these characteristics are, therefore, not defined. In this work, we built a model to automatically predict whether short clinical statements were considered inclusion or exclusion criteria. We used protocols from cancer clinical trials that were available in public registries from the last 18 years to train word-embeddings, and we constructed a~dataset of 6M short free-texts labeled as eligible or not eligible. A text classifier was trained using deep neural networks, with pre-trained word-embeddings as inputs, to predict whether or not short free-text statements describing clinical information were considered eligible. We additionally analyzed the semantic reasoning of the word-embedding representations obtained and were able to identify equivalent treatments for a type of tumor analogous with the drugs used to treat other tumors. We show that representation learning using {deep} neural networks can be successfully leveraged to extract the medical knowledge from clinical trial protocols for potentially assisting practitioners when prescribing treatments.
MirBot: A collaborative object recognition system for smartphones using convolutional neural networks
Mar 24, 2018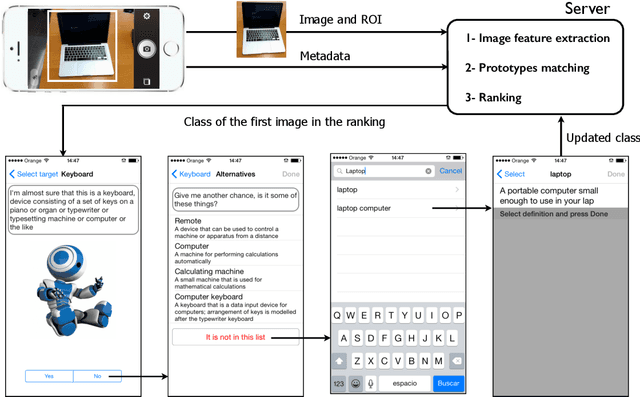
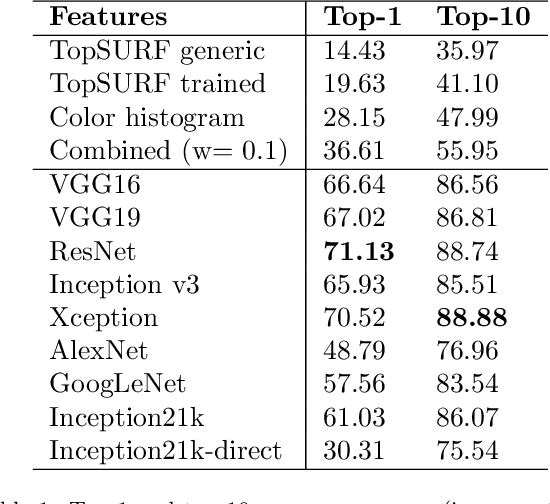
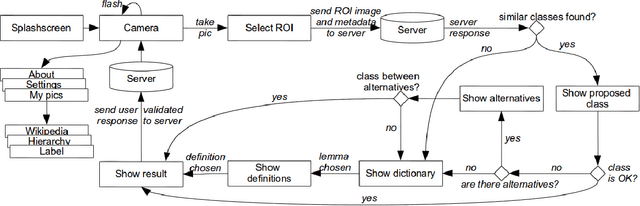
Abstract:MirBot is a collaborative application for smartphones that allows users to perform object recognition. This app can be used to take a photograph of an object, select the region of interest and obtain the most likely class (dog, chair, etc.) by means of similarity search using features extracted from a convolutional neural network (CNN). The answers provided by the system can be validated by the user so as to improve the results for future queries. All the images are stored together with a series of metadata, thus enabling a multimodal incremental dataset labeled with synset identifiers from the WordNet ontology. This dataset grows continuously thanks to the users' feedback, and is publicly available for research. This work details the MirBot object recognition system, analyzes the statistics gathered after more than four years of usage, describes the image classification methodology, and performs an exhaustive evaluation using handcrafted features, convolutional neural codes and different transfer learning techniques. After comparing various models and transformation methods, the results show that the CNN features maintain the accuracy of MirBot constant over time, despite the increasing number of new classes. The app is freely available at the Apple and Google Play stores.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge