Perminder S. Sachdev
for the Sydney Memory and Ageing Study and the Alzheimer's Disease Neuroimaging Initiative
Temporal Pattern Mining for Analysis of Longitudinal Clinical Data: Identifying Risk Factors for Alzheimer's Disease
Sep 11, 2022
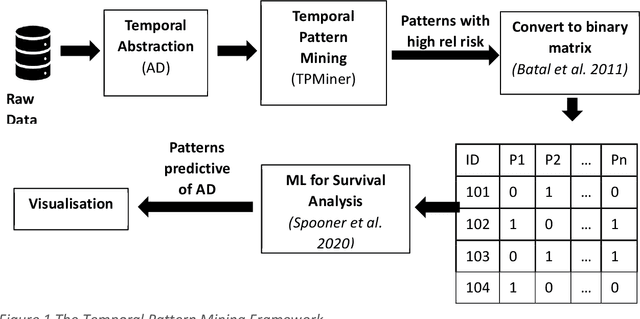

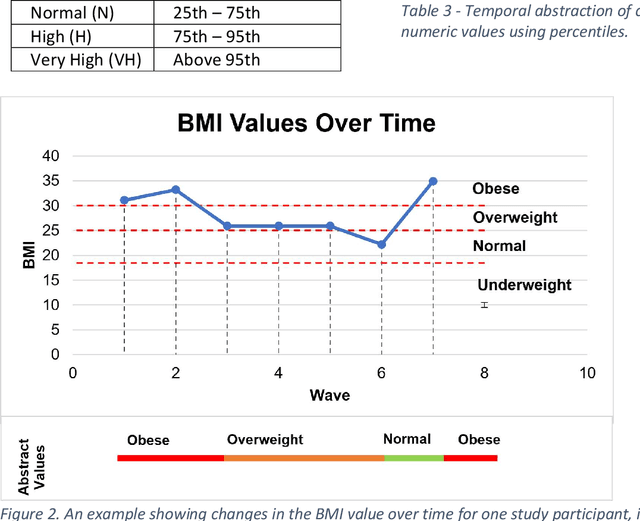
Abstract:A novel framework is proposed for handling the complex task of modelling and analysis of longitudinal, multivariate, heterogeneous clinical data. This method uses temporal abstraction to convert the data into a more appropriate form for modelling, temporal pattern mining, to discover patterns in the complex, longitudinal data and machine learning models of survival analysis to select the discovered patterns. The method is applied to a real-world study of Alzheimer's disease (AD), a progressive neurodegenerative disease that has no cure. The patterns discovered were predictive of AD in survival analysis models with a Concordance index of up to 0.8. This is the first work that performs survival analysis of AD data using temporal data collections for AD. A visualisation module also provides a clear picture of the discovered patterns for ease of interpretability.
Ensemble feature selection with clustering for analysis of high-dimensional, correlated clinical data in the search for Alzheimer's disease biomarkers
Jul 06, 2022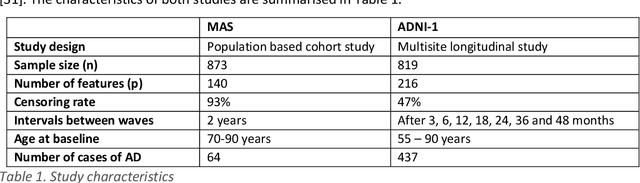
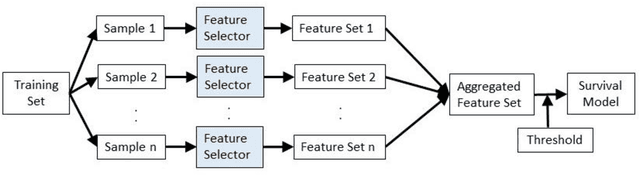
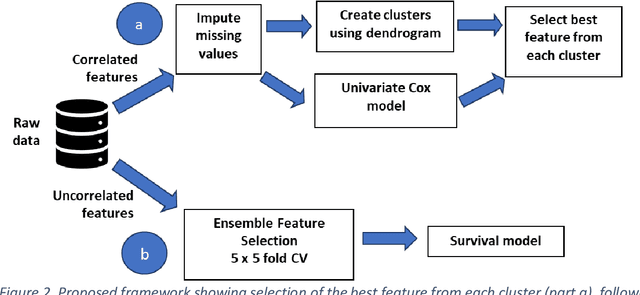
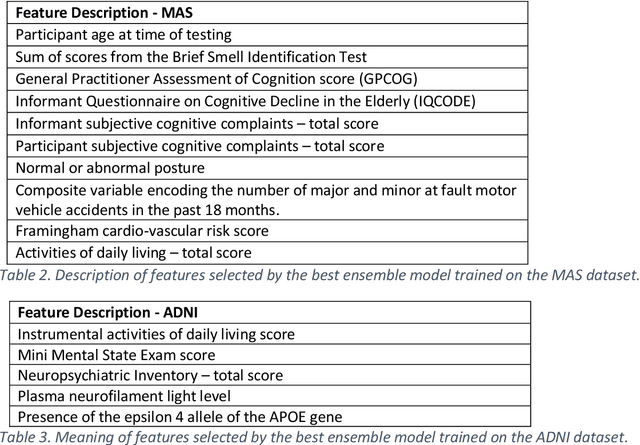
Abstract:Healthcare datasets often contain groups of highly correlated features, such as features from the same biological system. When feature selection is applied to these datasets to identify the most important features, the biases inherent in some multivariate feature selectors due to correlated features make it difficult for these methods to distinguish between the important and irrelevant features and the results of the feature selection process can be unstable. Feature selection ensembles, which aggregate the results of multiple individual base feature selectors, have been investigated as a means of stabilising feature selection results, but do not address the problem of correlated features. We present a novel framework to create feature selection ensembles from multivariate feature selectors while taking into account the biases produced by groups of correlated features, using agglomerative hierarchical clustering in a pre-processing step. These methods were applied to two real-world datasets from studies of Alzheimer's disease (AD), a progressive neurodegenerative disease that has no cure and is not yet fully understood. Our results show a marked improvement in the stability of features selected over the models without clustering, and the features selected by these models are in keeping with the findings in the AD literature.
Ensemble feature selection with data-driven thresholding for Alzheimer's disease biomarker discovery
Jul 05, 2022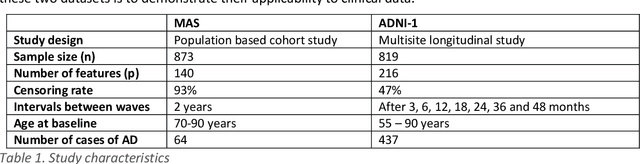

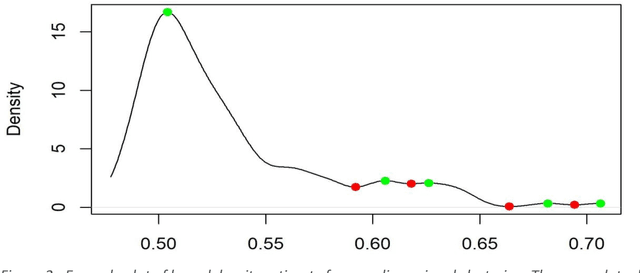
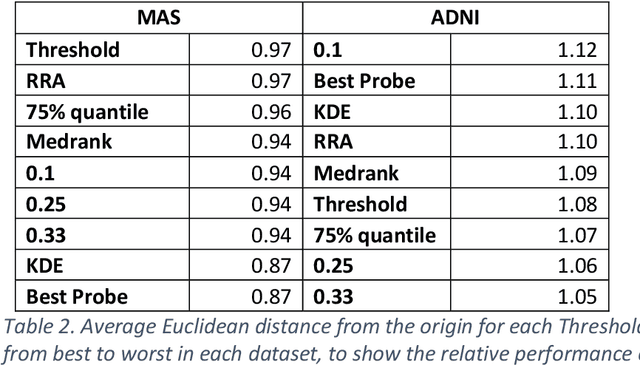
Abstract:Healthcare datasets present many challenges to both machine learning and statistics as their data are typically heterogeneous, censored, high-dimensional and have missing information. Feature selection is often used to identify the important features but can produce unstable results when applied to high-dimensional data, selecting a different set of features on each iteration. The stability of feature selection can be improved with the use of feature selection ensembles, which aggregate the results of multiple base feature selectors. A threshold must be applied to the final aggregated feature set to separate the relevant features from the redundant ones. A fixed threshold, which is typically applied, offers no guarantee that the final set of selected features contains only relevant features. This work develops several data-driven thresholds to automatically identify the relevant features in an ensemble feature selector and evaluates their predictive accuracy and stability. To demonstrate the applicability of these methods to clinical data, they are applied to data from two real-world Alzheimer's disease (AD) studies. AD is a progressive neurodegenerative disease with no known cure, that begins at least 2-3 decades before overt symptoms appear, presenting an opportunity for researchers to identify early biomarkers that might identify patients at risk of developing AD. Features identified by applying these methods to both datasets reflect current findings in the AD literature.
Computer-Aided Extraction of Select MRI Markers of Cerebral Small Vessel Disease: A Systematic Review
Apr 04, 2022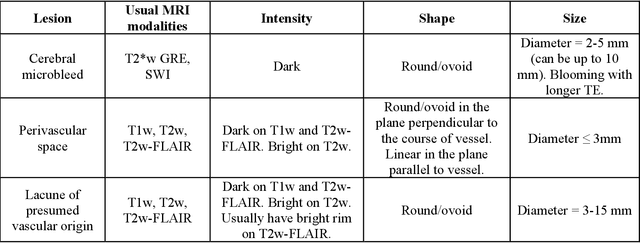
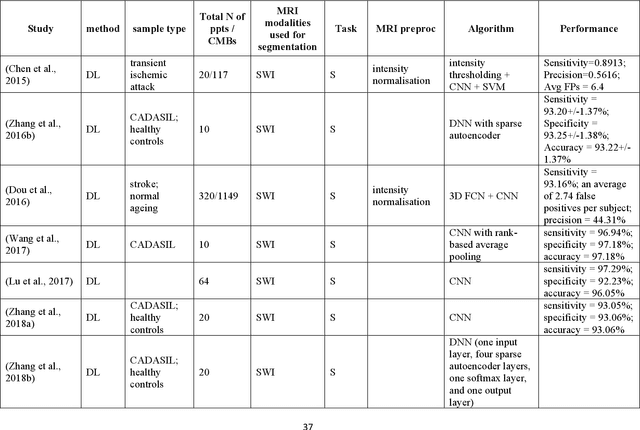
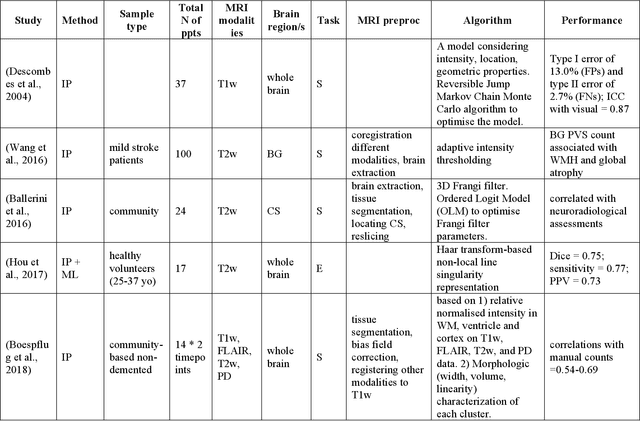
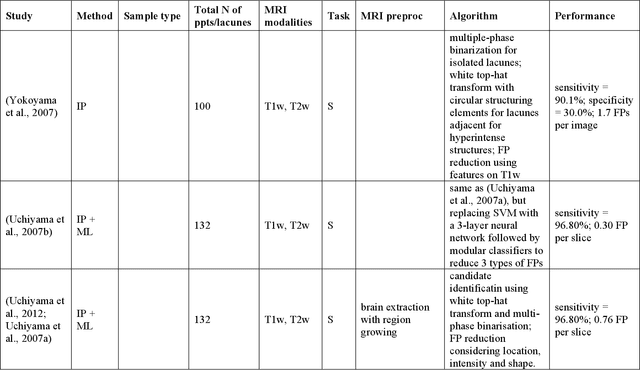
Abstract:Cerebral small vessel disease (CSVD) is a major vascular contributor to cognitive impairment in ageing, including dementias. Imaging remains the most promising method for in vivo studies of CSVD. To replace the subjective and laborious visual rating approaches, emerging studies have applied state-of-the-art artificial intelligence to extract imaging biomarkers of CSVD from MRI scans. We aimed to summarise published computer-aided methods to examine three imaging biomarkers of CSVD, namely cerebral microbleeds (CMB), dilated perivascular spaces (PVS), and lacunes of presumed vascular origin. Seventy-one classical image processing, classical machine learning, and deep learning studies were identified. CMB and PVS have been better studied, compared to lacunes. While good performance metrics have been achieved in local test datasets, there have not been generalisable pipelines validated in different research or clinical cohorts. Transfer learning and weak supervision techniques have been applied to accommodate the limitations in training data. Future studies could consider pooling data from multiple sources to increase diversity, and validating the performance of the methods using both image processing metrics and associations with clinical measures.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge