Ning Song
Chromosomal Structural Abnormality Diagnosis by Homologous Similarity
Jul 11, 2024Abstract:Pathogenic chromosome abnormalities are very common among the general population. While numerical chromosome abnormalities can be quickly and precisely detected, structural chromosome abnormalities are far more complex and typically require considerable efforts by human experts for identification. This paper focuses on investigating the modeling of chromosome features and the identification of chromosomes with structural abnormalities. Most existing data-driven methods concentrate on a single chromosome and consider each chromosome independently, overlooking the crucial aspect of homologous chromosomes. In normal cases, homologous chromosomes share identical structures, with the exception that one of them is abnormal. Therefore, we propose an adaptive method to align homologous chromosomes and diagnose structural abnormalities through homologous similarity. Inspired by the process of human expert diagnosis, we incorporate information from multiple pairs of homologous chromosomes simultaneously, aiming to reduce noise disturbance and improve prediction performance. Extensive experiments on real-world datasets validate the effectiveness of our model compared to baselines.
Varifocal-Net: A Chromosome Classification Approach using Deep Convolutional Networks
Oct 20, 2018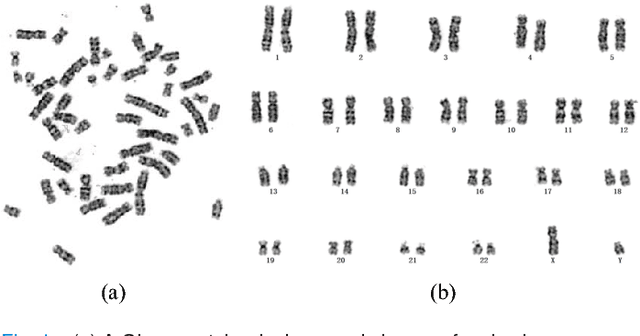
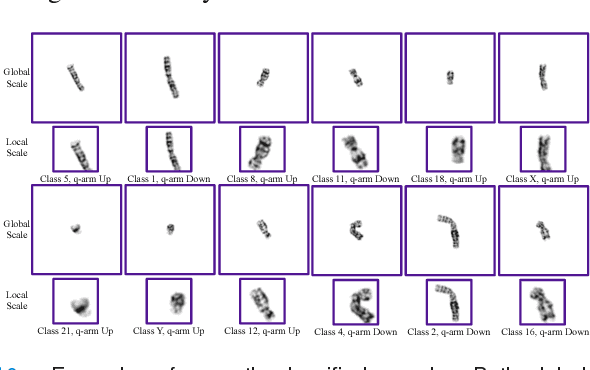
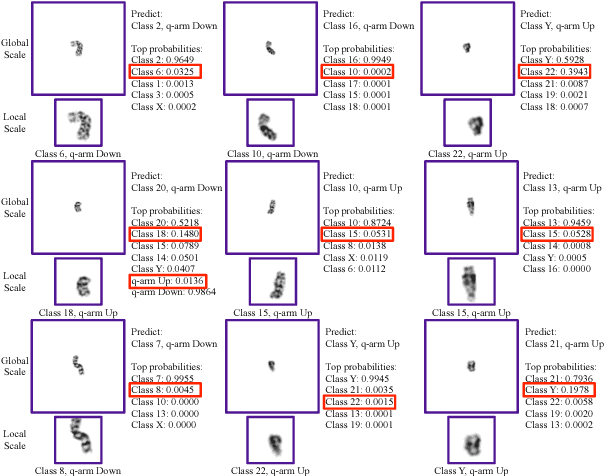
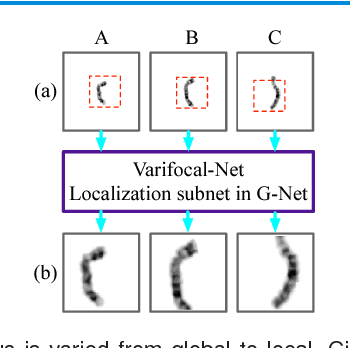
Abstract:Chromosome classification is critical for karyotyping in abnormality diagnosis. To expedite diagnosis process, we present a novel method named Varifocal-Net for simultaneous classification of chromosome's type and polarity using deep convolutional networks. The approach consists of one global-scale network (G-Net) and one local-scale network (L-Net). It follows two stages. The first stage is to learn both global and local features. We extract global features and detect finer local regions via the G-Net. With the proposed varifocal mechanism, we zoom into local parts and extract local features via the L-Net. Residual learning and multi-task learning strategies are utilized to promote high-level feature extraction. The detection of discriminative local parts is fulfilled by a localization subnet of the G-Net, whose training process involves both supervised and weekly-supervised learning. The second stage is to build two multi-layer perceptron classifiers that exploit features of both two scales to boost classification performance. Evaluation results from 1909 karyotyping cases demonstrate that our Varifocal-Net achieved the highest accuracy of 0.9805, 0.9909 and average F1-score of 0.9771, 0.9909 for the type and polarity task, respectively. It outperformed state-of-the-art methods, demonstrating the effectiveness of our Varifocal mechanism and multi-scale feature ensemble.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge