Fanzhe Fu
Aletheia: Quantifying Cognitive Conviction in Reasoning Models via Regularized Inverse Confusion Matrix
Jan 04, 2026Abstract:In the progressive journey toward Artificial General Intelligence (AGI), current evaluation paradigms face an epistemological crisis. Static benchmarks measure knowledge breadth but fail to quantify the depth of belief. While Simhi et al. (2025) defined the CHOKE phenomenon in standard QA, we extend this framework to quantify "Cognitive Conviction" in System 2 reasoning models. We propose Project Aletheia, a cognitive physics framework that employs Tikhonov Regularization to invert the judge's confusion matrix. To validate this methodology without relying on opaque private data, we implement a Synthetic Proxy Protocol. Our preliminary pilot study on 2025 baselines (e.g., DeepSeek-R1, OpenAI o1) suggests that while reasoning models act as a "cognitive buffer," they may exhibit "Defensive OverThinking" under adversarial pressure. Furthermore, we introduce the Aligned Conviction Score (S_aligned) to verify that conviction does not compromise safety. This work serves as a blueprint for measuring AI scientific integrity.
The Meta-Prompting Protocol: Orchestrating LLMs via Adversarial Feedback Loops
Dec 17, 2025

Abstract:The transition of Large Language Models (LLMs) from stochastic chat interfaces to reliable software components necessitates a fundamental re-engineering of interaction paradigms. Current methodologies, predominantly heuristic-based "prompt engineering," fail to provide the deterministic guarantees required for mission-critical applications. We introduce the Meta-Prompting Protocol, a rigorous theoretical framework that formalizes the orchestration of LLMs as a programmable, self-optimizing system. Central to this protocol is the Adversarial Trinity, a tripartite topology comprising a Generator (P), an Auditor (A), and an Optimizer (O). By treating natural language instructions as differentiable variables within a semantic computation graph and utilizing textual critiques as gradients, this architecture mitigates hallucination and prevents model collapse. We demonstrate the theoretical viability of this approach using declarative programming paradigms (DSPy) and automatic textual differentiation (TextGrad), establishing a foundation for "Observable Software Engineering" in the era of probabilistic computing.
Noise or Signal? Deconstructing Contradictions and An Adaptive Remedy for Reversible Normalization in Time Series Forecasting
Oct 06, 2025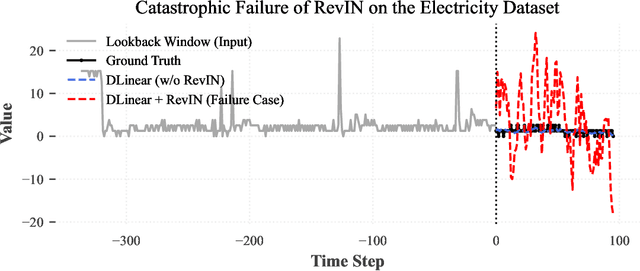
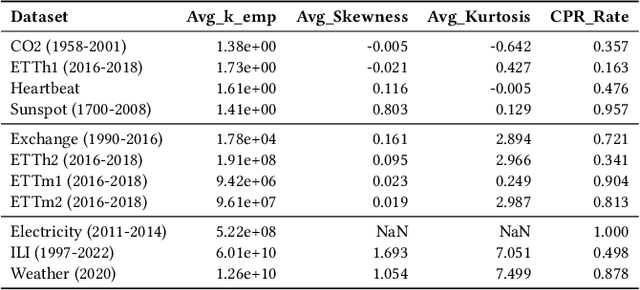
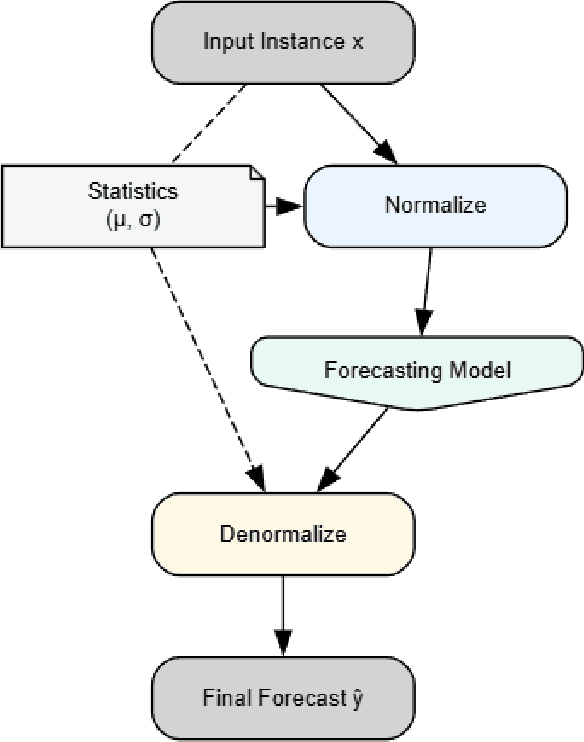
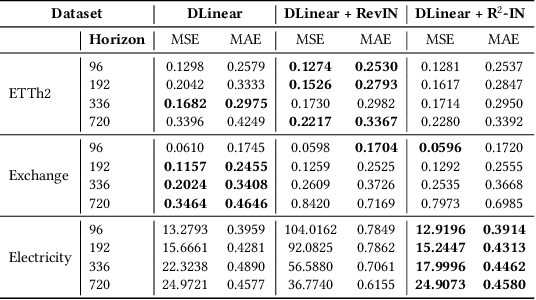
Abstract:Reversible Instance Normalization (RevIN) is a key technique enabling simple linear models to achieve state-of-the-art performance in time series forecasting. While replacing its non-robust statistics with robust counterparts (termed R$^2$-IN) seems like a straightforward improvement, our findings reveal a far more complex reality. This paper deconstructs the perplexing performance of various normalization strategies by identifying four underlying theoretical contradictions. Our experiments provide two crucial findings: first, the standard RevIN catastrophically fails on datasets with extreme outliers, where its MSE surges by a staggering 683\%. Second, while the simple R$^2$-IN prevents this failure and unexpectedly emerges as the best overall performer, our adaptive model (A-IN), designed to test a diagnostics-driven heuristic, unexpectedly suffers a complete and systemic failure. This surprising outcome uncovers a critical, overlooked pitfall in time series analysis: the instability introduced by a simple or counter-intuitive heuristic can be more damaging than the statistical issues it aims to solve. The core contribution of this work is thus a new, cautionary paradigm for time series normalization: a shift from a blind search for complexity to a diagnostics-driven analysis that reveals not only the surprising power of simple baselines but also the perilous nature of naive adaptation.
Chromosomal Structural Abnormality Diagnosis by Homologous Similarity
Jul 11, 2024Abstract:Pathogenic chromosome abnormalities are very common among the general population. While numerical chromosome abnormalities can be quickly and precisely detected, structural chromosome abnormalities are far more complex and typically require considerable efforts by human experts for identification. This paper focuses on investigating the modeling of chromosome features and the identification of chromosomes with structural abnormalities. Most existing data-driven methods concentrate on a single chromosome and consider each chromosome independently, overlooking the crucial aspect of homologous chromosomes. In normal cases, homologous chromosomes share identical structures, with the exception that one of them is abnormal. Therefore, we propose an adaptive method to align homologous chromosomes and diagnose structural abnormalities through homologous similarity. Inspired by the process of human expert diagnosis, we incorporate information from multiple pairs of homologous chromosomes simultaneously, aiming to reduce noise disturbance and improve prediction performance. Extensive experiments on real-world datasets validate the effectiveness of our model compared to baselines.
Are Synthetic Time-series Data Really not as Good as Real Data?
Feb 01, 2024Abstract:Time-series data presents limitations stemming from data quality issues, bias and vulnerabilities, and generalization problem. Integrating universal data synthesis methods holds promise in improving generalization. However, current methods cannot guarantee that the generator's output covers all unseen real data. In this paper, we introduce InfoBoost -- a highly versatile cross-domain data synthesizing framework with time series representation learning capability. We have developed a method based on synthetic data that enables model training without the need for real data, surpassing the performance of models trained with real data. Additionally, we have trained a universal feature extractor based on our synthetic data that is applicable to all time-series data. Our approach overcomes interference from multiple sources rhythmic signal, noise interference, and long-period features that exceed sampling window capabilities. Through experiments, our non-deep-learning synthetic data enables models to achieve superior reconstruction performance and universal explicit representation extraction without the need for real data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge