Nico Pfeifer
Privacy Preserving Federated Unsupervised Domain Adaptation with Application to Age Prediction from DNA Methylation Data
Nov 26, 2024Abstract:In computational biology, predictive models are widely used to address complex tasks, but their performance can suffer greatly when applied to data from different distributions. The current state-of-the-art domain adaptation method for high-dimensional data aims to mitigate these issues by aligning the input dependencies between training and test data. However, this approach requires centralized access to both source and target domain data, raising concerns about data privacy, especially when the data comes from multiple sources. In this paper, we introduce a privacy-preserving federated framework for unsupervised domain adaptation in high-dimensional settings. Our method employs federated training of Gaussian processes and weighted elastic nets to effectively address the problem of distribution shift between domains, while utilizing secure aggregation and randomized encoding to protect the local data of participating data owners. We evaluate our framework on the task of age prediction using DNA methylation data from multiple tissues, demonstrating that our approach performs comparably to existing centralized methods while maintaining data privacy, even in distributed environments where data is spread across multiple institutions. Our framework is the first privacy-preserving solution for high-dimensional domain adaptation in federated environments, offering a promising tool for fields like computational biology and medicine, where protecting sensitive data is essential.
Robust Representation Learning for Privacy-Preserving Machine Learning: A Multi-Objective Autoencoder Approach
Sep 08, 2023Abstract:Several domains increasingly rely on machine learning in their applications. The resulting heavy dependence on data has led to the emergence of various laws and regulations around data ethics and privacy and growing awareness of the need for privacy-preserving machine learning (ppML). Current ppML techniques utilize methods that are either purely based on cryptography, such as homomorphic encryption, or that introduce noise into the input, such as differential privacy. The main criticism given to those techniques is the fact that they either are too slow or they trade off a model s performance for improved confidentiality. To address this performance reduction, we aim to leverage robust representation learning as a way of encoding our data while optimizing the privacy-utility trade-off. Our method centers on training autoencoders in a multi-objective manner and then concatenating the latent and learned features from the encoding part as the encoded form of our data. Such a deep learning-powered encoding can then safely be sent to a third party for intensive training and hyperparameter tuning. With our proposed framework, we can share our data and use third party tools without being under the threat of revealing its original form. We empirically validate our results on unimodal and multimodal settings, the latter following a vertical splitting system and show improved performance over state-of-the-art.
PlasmoFAB: A Benchmark to Foster Machine Learning for Plasmodium falciparum Protein Antigen Candidate Prediction
Jan 16, 2023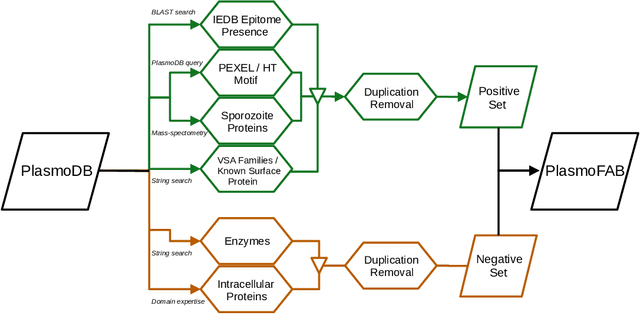
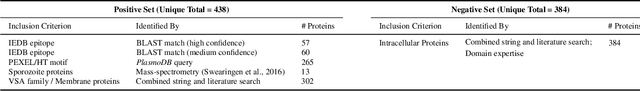
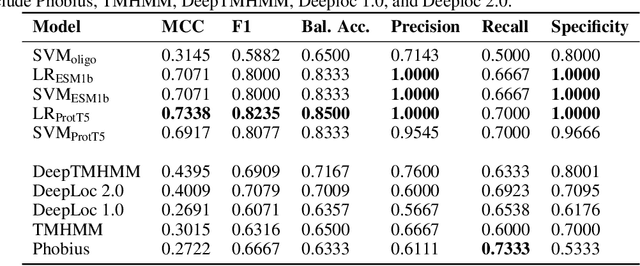
Abstract:Motivation: Machine learning methods can be used to support scientific discovery in healthcare-related research fields. However, these methods can only be reliably used if they can be trained on high-quality and curated datasets. Currently, no such dataset for the exploration of Plasmodium falciparum protein antigen candidates exists. The parasite Plasmodium falciparum causes the infectious disease malaria. Thus, identifying potential antigens is of utmost importance for the development of antimalarial drugs and vaccines. Since exploring antigen candidates experimentally is an expensive and time-consuming process, applying machine learning methods to support this process has the potential to accelerate the development of drugs and vaccines which are needed for fighting and controlling malaria. Results: We developed PlasmoFAB, a curated benchmark that can be used to train machine learning methods for the exploration of Plasmodium falciparum protein antigen candidates. We combined an extensive literature search with domain expertise to create high-quality labels for Plasmodium falciparum specific proteins that distinguish between antigen candidates and intracellular proteins. Additionally, we used our benchmark to compare different well-known prediction models and available protein localization prediction services on the task of identifying protein antigen candidates. We show that available general-purpose services are unable to provide sufficient performance on identifying protein antigen candidates and are outperformed by models that were trained on specialized data.
Bringing the Algorithms to the Data -- Secure Distributed Medical Analytics using the Personal Health Train
Dec 07, 2022Abstract:The need for data privacy and security -- enforced through increasingly strict data protection regulations -- renders the use of healthcare data for machine learning difficult. In particular, the transfer of data between different hospitals is often not permissible and thus cross-site pooling of data not an option. The Personal Health Train (PHT) paradigm proposed within the GO-FAIR initiative implements an 'algorithm to the data' paradigm that ensures that distributed data can be accessed for analysis without transferring any sensitive data. We present PHT-meDIC, a productively deployed open-source implementation of the PHT concept. Containerization allows us to easily deploy even complex data analysis pipelines (e.g, genomics, image analysis) across multiple sites in a secure and scalable manner. We discuss the underlying technological concepts, security models, and governance processes. The implementation has been successfully applied to distributed analyses of large-scale data, including applications of deep neural networks to medical image data.
COmic: Convolutional Kernel Networks for Interpretable End-to-End Learning on (Multi-)Omics Data
Dec 02, 2022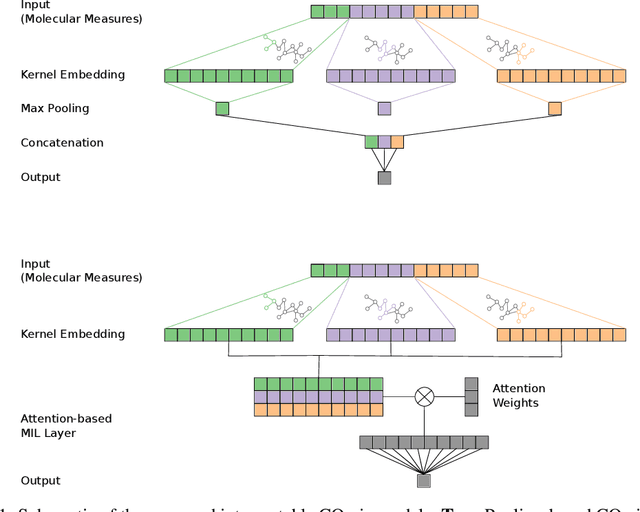
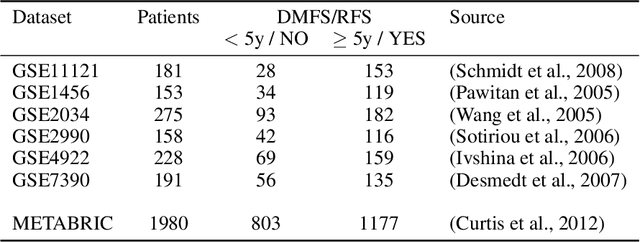
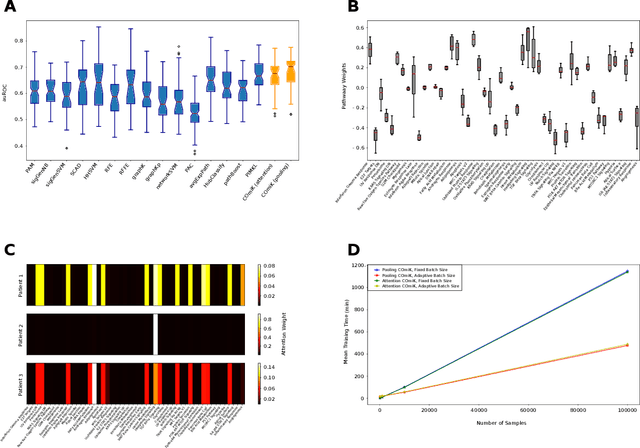
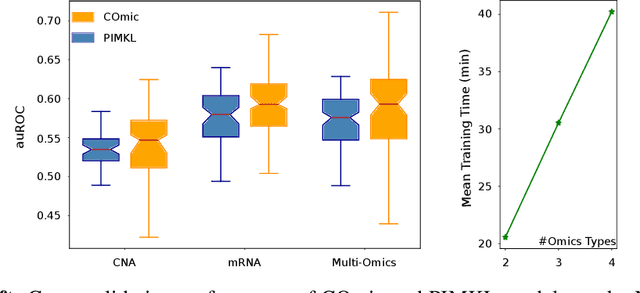
Abstract:Motivation: The size of available omics datasets is steadily increasing with technological advancement in recent years. While this increase in sample size can be used to improve the performance of relevant prediction tasks in healthcare, models that are optimized for large datasets usually operate as black boxes. In high stakes scenarios, like healthcare, using a black-box model poses safety and security issues. Without an explanation about molecular factors and phenotypes that affected the prediction, healthcare providers are left with no choice but to blindly trust the models. We propose a new type of artificial neural networks, named Convolutional Omics Kernel Networks (COmic). By combining convolutional kernel networks with pathway-induced kernels, our method enables robust and interpretable end-to-end learning on omics datasets ranging in size from a few hundred to several hundreds of thousands of samples. Furthermore, COmic can be easily adapted to utilize multi-omics data. Results: We evaluate the performance capabilities of COmic on six different breast cancer cohorts. Additionally, we train COmic models on multi-omics data using the METABRIC cohort. Our models perform either better or similar to competitors on both tasks. We show how the use of pathway-induced Laplacian kernels opens the black-box nature of neural networks and results in intrinsically interpretable models that eliminate the need for \textit{post-hoc} explanation models.
CECILIA: Comprehensive Secure Machine Learning Framework
Feb 07, 2022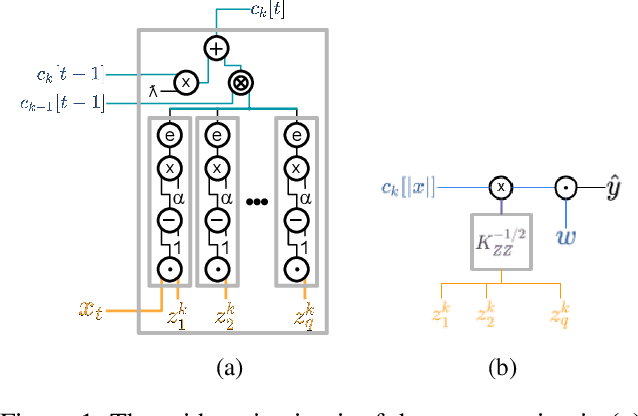
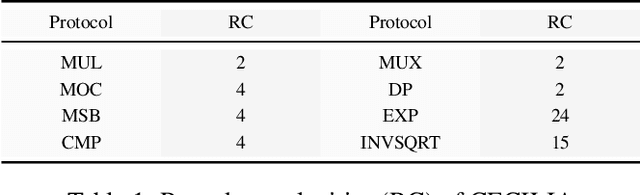
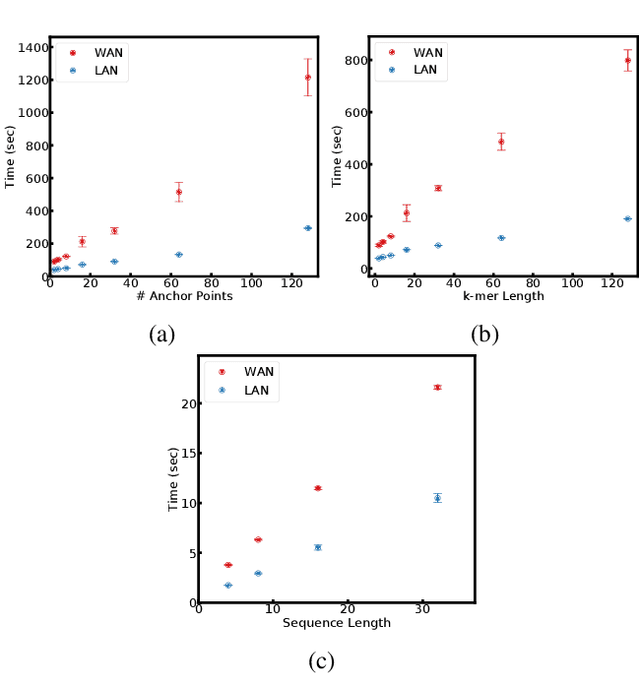
Abstract:Since machine learning algorithms have proven their success in data mining tasks, the data with sensitive information enforce privacy preserving machine learning algorithms to emerge. Moreover, the increase in the number of data sources and the high computational power required by those algorithms force individuals to outsource the training and/or the inference of a machine learning model to the clouds providing such services. To address this dilemma, we propose a secure 3-party computation framework, CECILIA, offering privacy preserving building blocks to enable more complex operations privately. Among those building blocks, we have two novel methods, which are the exact exponential of a public base raised to the power of a secret value and the inverse square root of a secret Gram matrix. We employ CECILIA to realize the private inference on pre-trained recurrent kernel networks, which require more complex operations than other deep neural networks such as convolutional neural networks, on the structural classification of proteins as the first study ever accomplishing the privacy preserving inference on recurrent kernel networks. The results demonstrate that we perform the exact and fully private exponential computation, which is done by approximation in the literature so far. Moreover, we can also perform the exact inverse square root of a secret Gram matrix computation up to a certain privacy level, which has not been addressed in the literature at all. We also analyze the scalability of CECILIA to various settings on a synthetic dataset. The framework shows a great promise to make other machine learning algorithms as well as further computations privately computable by the building blocks of the framework.
Convolutional Motif Kernel Networks
Nov 03, 2021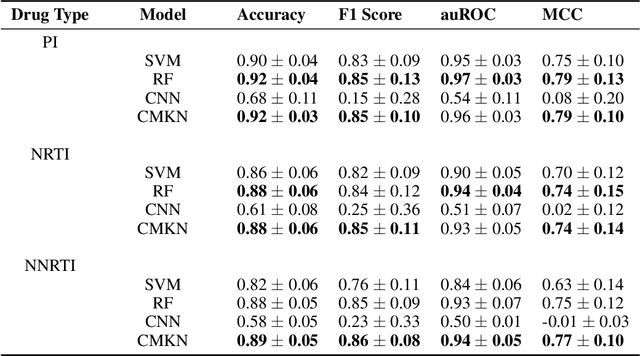
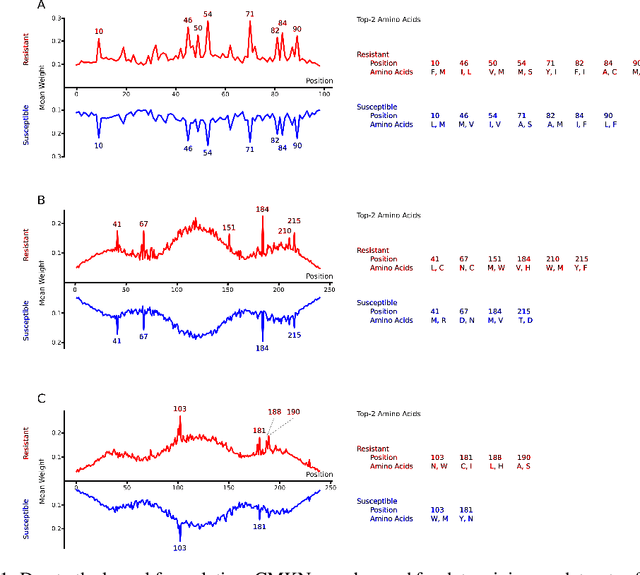
Abstract:Artificial neural networks are exceptionally good in learning to detect correlations within data that are associated with specified outcomes. However to deepen knowledge and support further research, researchers have to be able to explain predicted outcomes within the data's domain. Furthermore, domain experts like Healthcare Providers need these explanations to assess whether a predicted outcome can be trusted in high stakes scenarios and to help them incorporating a model into their own routine. In this paper we introduce Convolutional Motif Kernel Networks, a neural network architecture that incorporates learning a feature representation within a subspace of the reproducing kernel Hilbert space of the motif kernel function. The resulting model has state-of-the-art performance and enables researchers and domain experts to directly interpret and verify prediction outcomes without the need for a post hoc explainability method.
ppAUC: Privacy Preserving Area Under the Curve with Secure 3-Party Computation
Feb 17, 2021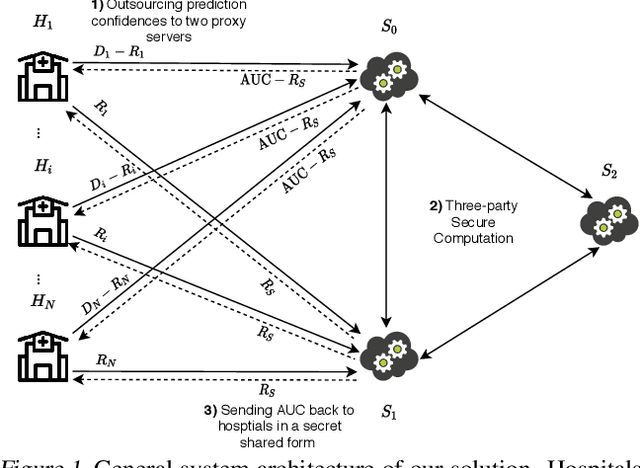
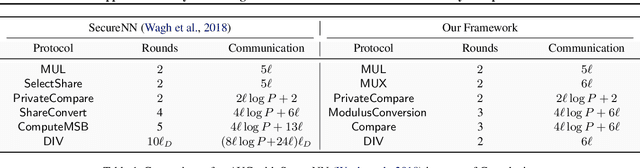
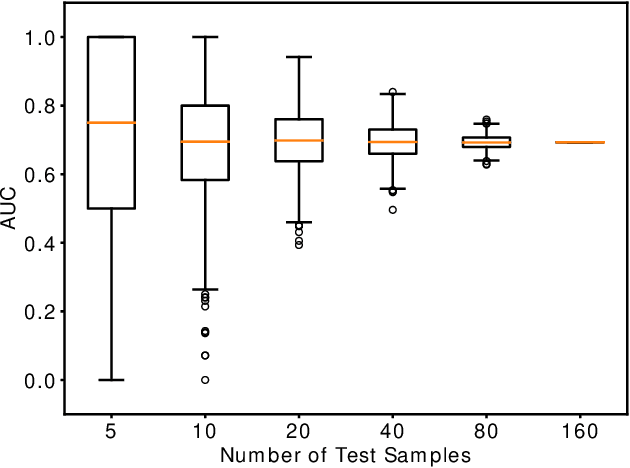
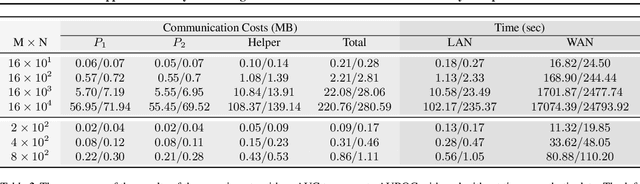
Abstract:Computing an AUC as a performance measure to compare the quality of different machine learning models is one of the final steps of many research projects. Many of these methods are trained on privacy-sensitive data and there are several different approaches like $\epsilon$-differential privacy, federated machine learning and methods based on cryptographic approaches if the datasets cannot be shared or evaluated jointly at one place. In this setting, it can also be a problem to compute the global AUC, since the labels might also contain privacy-sensitive information. There have been approaches based on $\epsilon$-differential privacy to deal with this problem, but to the best of our knowledge, no exact privacy preserving solution has been introduced. In this paper, we propose an MPC-based framework, called privacy preserving AUC (ppAUC), with novel methods for comparing two secret-shared values, selecting between two secret-shared values, converting the modulus and performing division to compute the exact AUC as one could obtain on the pooled original test samples. We employ ppAUC in the computation of the exact area under precision-recall curve and receiver operating characteristic curve even for ties between prediction confidence values. To prove the correctness of ppAUC, we apply it to evaluate a model trained to predict acute myeloid leukemia therapy response and we also assess its scalability via experiments on synthetic data. The experiments show that we efficiently compute exactly the same AUC with both evaluation metrics in a privacy preserving manner as one can obtain on the pooled test samples in the plaintext domain. Our solution provides security against semi-honest corruption of at most one of the servers performing the secure computation.
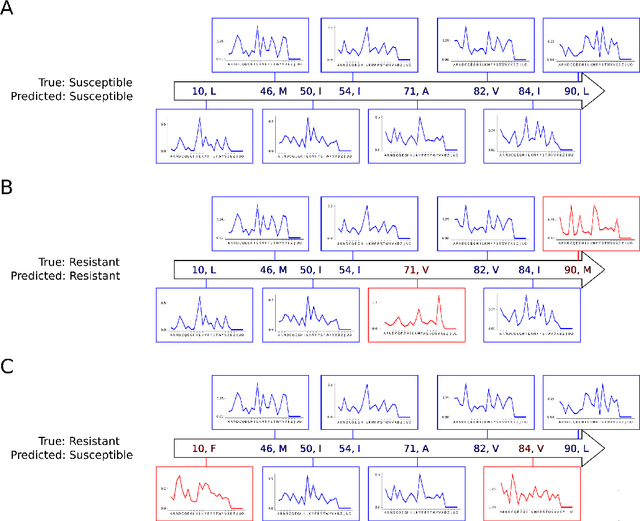
ESCAPED: Efficient Secure and Private Dot Product Framework for Kernel-based Machine Learning Algorithms with Applications in Healthcare
Dec 04, 2020
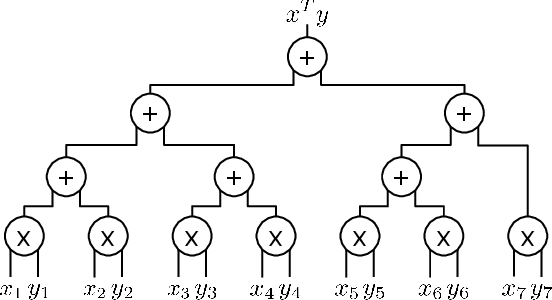
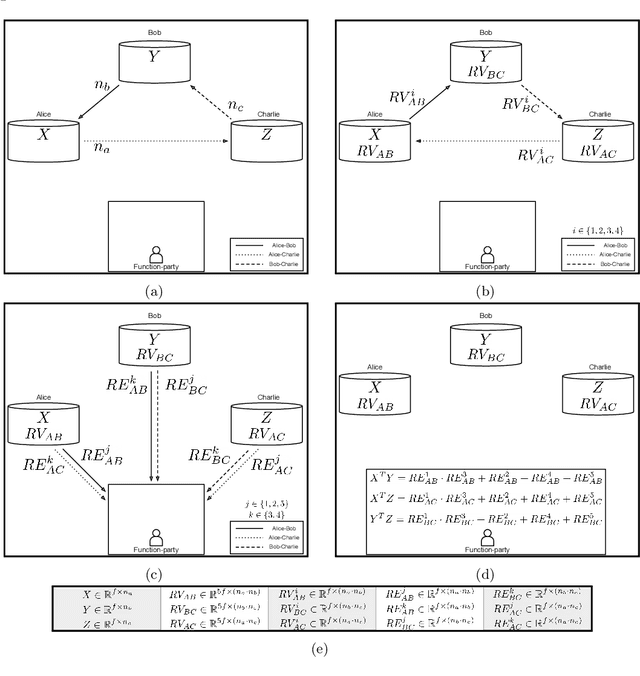
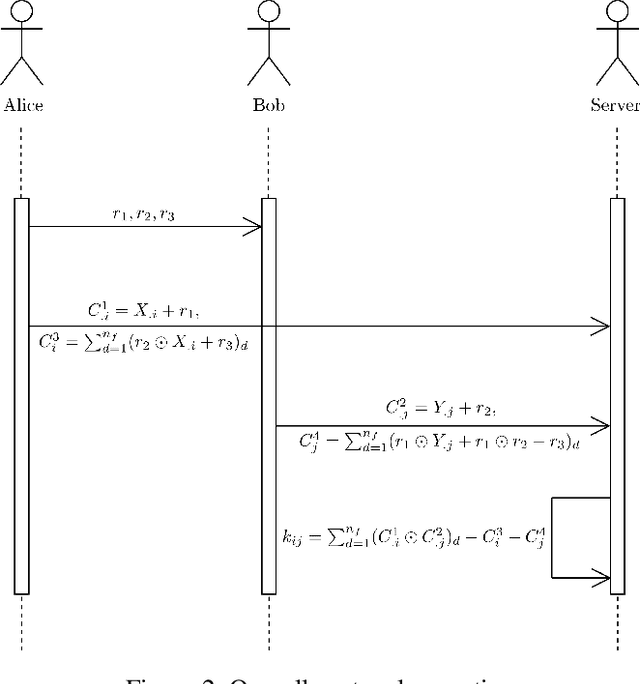
Abstract:To train sophisticated machine learning models one usually needs many training samples. Especially in healthcare settings these samples can be very expensive, meaning that one institution alone usually does not have enough on its own. Merging privacy-sensitive data from different sources is usually restricted by data security and data protection measures. This can lead to approaches that reduce data quality by putting noise onto the variables (e.g., in $\epsilon$-differential privacy) or omitting certain values (e.g., for $k$-anonymity). Other measures based on cryptographic methods can lead to very time-consuming computations, which is especially problematic for larger multi-omics data. We address this problem by introducing ESCAPED, which stands for Efficient SeCure And PrivatE Dot product framework, enabling the computation of the dot product of vectors from multiple sources on a third-party, which later trains kernel-based machine learning algorithms, while neither sacrificing privacy nor adding noise. We evaluated our framework on drug resistance prediction for HIV-infected people and multi-omics dimensionality reduction and clustering problems in precision medicine. In terms of execution time, our framework significantly outperforms the best-fitting existing approaches without sacrificing the performance of the algorithm. Even though we only show the benefit for kernel-based algorithms, our framework can open up new research opportunities for further machine learning models that require the dot product of vectors from multiple sources.
Privacy Preserving Gaze Estimation using Synthetic Images via a Randomized Encoding Based Framework
Nov 06, 2019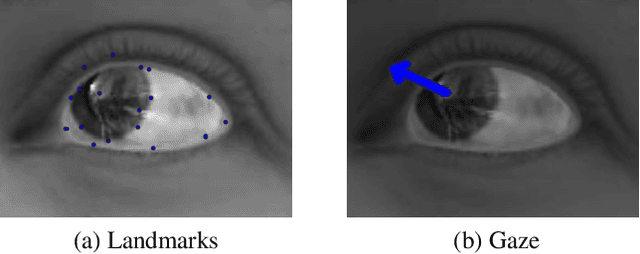
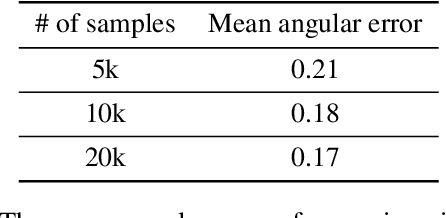
Abstract:Eye tracking is handled as one of the key technologies for applications which assess and evaluate human attention, behavior and biometrics, especially using gaze, pupillary and blink behaviors. One of the main challenges with regard to the social acceptance of eye-tracking technology is however the preserving of sensitive and personal information. To tackle this challenge, we employed a privacy-preserving framework based on randomized encoding to train a Support Vector Regression model on synthetic eye images privately to estimate human gaze. During the computation, none of the parties learns about the data or the result that any other party has. Furthermore, the party that trains the model cannot reconstruct pupil, blink or visual scanpath. The experimental results showed that our privacy preserving framework is also capable of working in real-time, as accurate as a non-private version of it and could be extended to other eye-tracking related problems.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge