Neda Zamanitajeddin
Benchmarking Domain Generalization Algorithms in Computational Pathology
Sep 25, 2024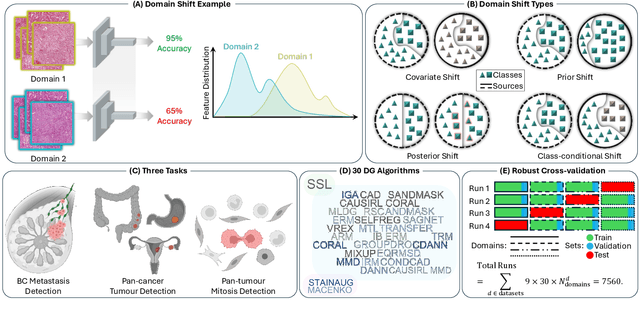
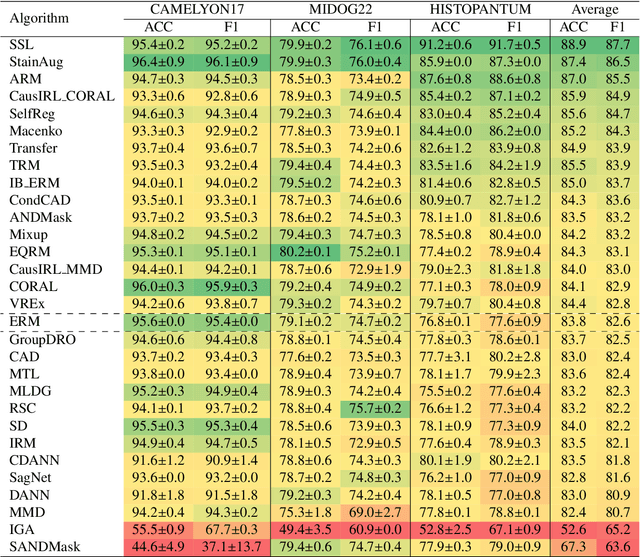
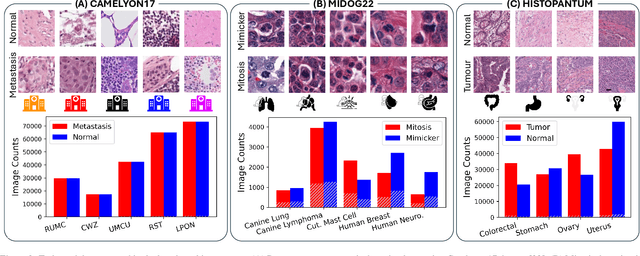
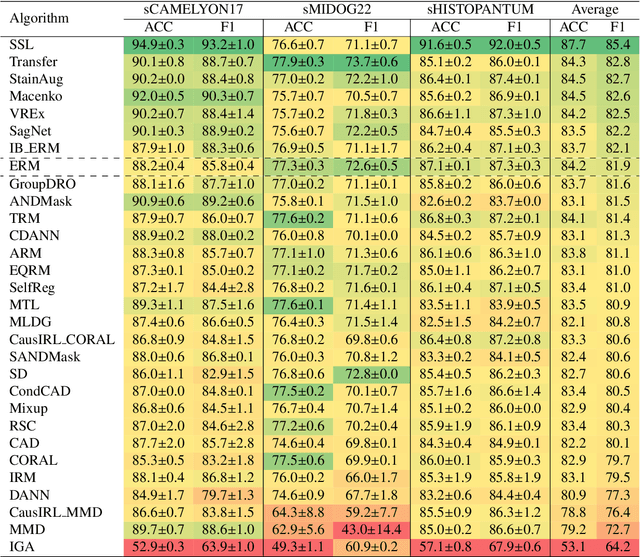
Abstract:Deep learning models have shown immense promise in computational pathology (CPath) tasks, but their performance often suffers when applied to unseen data due to domain shifts. Addressing this requires domain generalization (DG) algorithms. However, a systematic evaluation of DG algorithms in the CPath context is lacking. This study aims to benchmark the effectiveness of 30 DG algorithms on 3 CPath tasks of varying difficulty through 7,560 cross-validation runs. We evaluate these algorithms using a unified and robust platform, incorporating modality-specific techniques and recent advances like pretrained foundation models. Our extensive cross-validation experiments provide insights into the relative performance of various DG strategies. We observe that self-supervised learning and stain augmentation consistently outperform other methods, highlighting the potential of pretrained models and data augmentation. Furthermore, we introduce a new pan-cancer tumor detection dataset (HISTOPANTUM) as a benchmark for future research. This study offers valuable guidance to researchers in selecting appropriate DG approaches for CPath tasks.
Domain Generalization in Computational Pathology: Survey and Guidelines
Oct 30, 2023



Abstract:Deep learning models have exhibited exceptional effectiveness in Computational Pathology (CPath) by tackling intricate tasks across an array of histology image analysis applications. Nevertheless, the presence of out-of-distribution data (stemming from a multitude of sources such as disparate imaging devices and diverse tissue preparation methods) can cause \emph{domain shift} (DS). DS decreases the generalization of trained models to unseen datasets with slightly different data distributions, prompting the need for innovative \emph{domain generalization} (DG) solutions. Recognizing the potential of DG methods to significantly influence diagnostic and prognostic models in cancer studies and clinical practice, we present this survey along with guidelines on achieving DG in CPath. We rigorously define various DS types, systematically review and categorize existing DG approaches and resources in CPath, and provide insights into their advantages, limitations, and applicability. We also conduct thorough benchmarking experiments with 28 cutting-edge DG algorithms to address a complex DG problem. Our findings suggest that careful experiment design and CPath-specific Stain Augmentation technique can be very effective. However, there is no one-size-fits-all solution for DG in CPath. Therefore, we establish clear guidelines for detecting and managing DS depending on different scenarios. While most of the concepts, guidelines, and recommendations are given for applications in CPath, we believe that they are applicable to most medical image analysis tasks as well.
Stain-Robust Mitotic Figure Detection for MIDOG 2022 Challenge
Aug 26, 2022

Abstract:The detection of mitotic figures from different scanners/sites remains an important topic of research, owing to its potential in assisting clinicians with tumour grading. The MItosis DOmain Generalization (MIDOG) 2022 challenge aims to test the robustness of detection models on unseen data from multiple scanners and tissue types for this task. We present a short summary of the approach employed by the TIA Centre team to address this challenge. Our approach is based on a hybrid detection model, where mitotic candidates are segmented, before being refined by a deep learning classifier. Cross-validation on the training images achieved the F1-score of 0.816 and 0.784 on the preliminary test set, demonstrating the generalizability of our model to unseen data from new scanners.
Cells are Actors: Social Network Analysis with Classical ML for SOTA Histology Image Classification
Jul 10, 2021



Abstract:Digitization of histology images and the advent of new computational methods, like deep learning, have helped the automatic grading of colorectal adenocarcinoma cancer (CRA). Present automated CRA grading methods, however, usually use tiny image patches and thus fail to integrate the entire tissue micro-architecture for grading purposes. To tackle these challenges, we propose to use a statistical network analysis method to describe the complex structure of the tissue micro-environment by modelling nuclei and their connections as a network. We show that by analyzing only the interactions between the cells in a network, we can extract highly discriminative statistical features for CRA grading. Unlike other deep learning or convolutional graph-based approaches, our method is highly scalable (can be used for cell networks consist of millions of nodes), completely explainable, and computationally inexpensive. We create cell networks on a broad CRC histology image dataset, experiment with our method, and report state-of-the-art performance for the prediction of three-class CRA grading.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge