Rob Jewsbury
Domain Generalization in Computational Pathology: Survey and Guidelines
Oct 30, 2023



Abstract:Deep learning models have exhibited exceptional effectiveness in Computational Pathology (CPath) by tackling intricate tasks across an array of histology image analysis applications. Nevertheless, the presence of out-of-distribution data (stemming from a multitude of sources such as disparate imaging devices and diverse tissue preparation methods) can cause \emph{domain shift} (DS). DS decreases the generalization of trained models to unseen datasets with slightly different data distributions, prompting the need for innovative \emph{domain generalization} (DG) solutions. Recognizing the potential of DG methods to significantly influence diagnostic and prognostic models in cancer studies and clinical practice, we present this survey along with guidelines on achieving DG in CPath. We rigorously define various DS types, systematically review and categorize existing DG approaches and resources in CPath, and provide insights into their advantages, limitations, and applicability. We also conduct thorough benchmarking experiments with 28 cutting-edge DG algorithms to address a complex DG problem. Our findings suggest that careful experiment design and CPath-specific Stain Augmentation technique can be very effective. However, there is no one-size-fits-all solution for DG in CPath. Therefore, we establish clear guidelines for detecting and managing DS depending on different scenarios. While most of the concepts, guidelines, and recommendations are given for applications in CPath, we believe that they are applicable to most medical image analysis tasks as well.
A QuadTree Image Representation for Computational Pathology
Aug 24, 2021
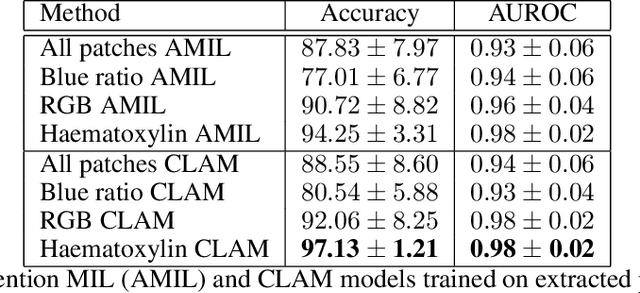

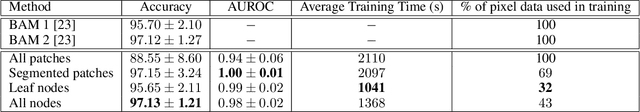
Abstract:The field of computational pathology presents many challenges for computer vision algorithms due to the sheer size of pathology images. Histopathology images are large and need to be split up into image tiles or patches so modern convolutional neural networks (CNNs) can process them. In this work, we present a method to generate an interpretable image representation of computational pathology images using quadtrees and a pipeline to use these representations for highly accurate downstream classification. To the best of our knowledge, this is the first attempt to use quadtrees for pathology image data. We show it is highly accurate, able to achieve as good results as the currently widely adopted tissue mask patch extraction methods all while using over 38% less data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge