Mostafa Karimi
Deploying self-supervised learning in the wild for hybrid automatic speech recognition
May 17, 2022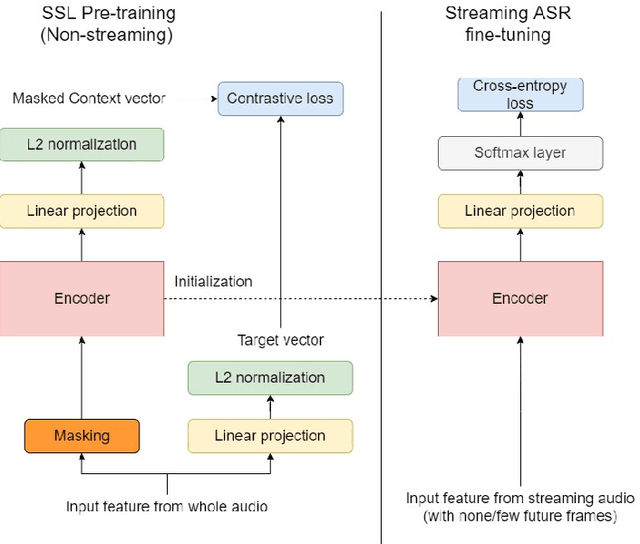
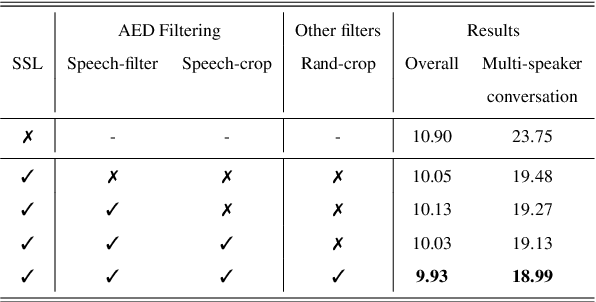
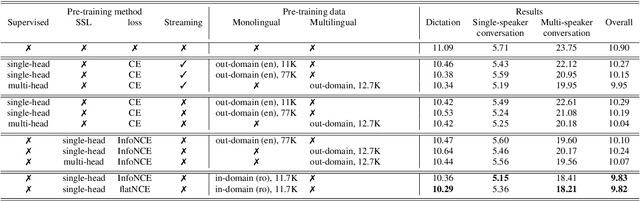
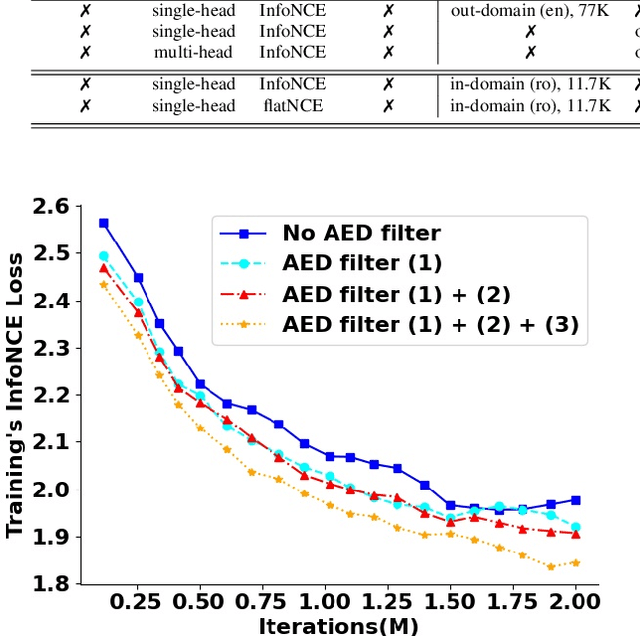
Abstract:Self-supervised learning (SSL) methods have proven to be very successful in automatic speech recognition (ASR). These great improvements have been reported mostly based on highly curated datasets such as LibriSpeech for non-streaming End-to-End ASR models. However, the pivotal characteristics of SSL is to be utilized for any untranscribed audio data. In this paper, we provide a full exploration on how to utilize uncurated audio data in SSL from data pre-processing to deploying an streaming hybrid ASR model. More specifically, we present (1) the effect of Audio Event Detection (AED) model in data pre-processing pipeline (2) analysis on choosing optimizer and learning rate scheduling (3) comparison of recently developed contrastive losses, (4) comparison of various pre-training strategies such as utilization of in-domain versus out-domain pre-training data, monolingual versus multilingual pre-training data, multi-head multilingual SSL versus single-head multilingual SSL and supervised pre-training versus SSL. The experimental results show that SSL pre-training with in-domain uncurated data can achieve better performance in comparison to all the alternative out-domain pre-training strategies.
Network-principled deep generative models for designing drug combinations as graph sets
Apr 22, 2020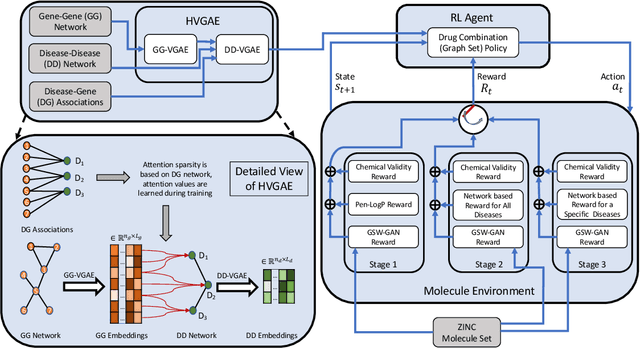
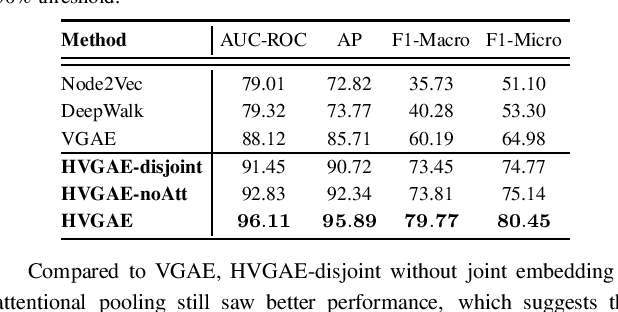
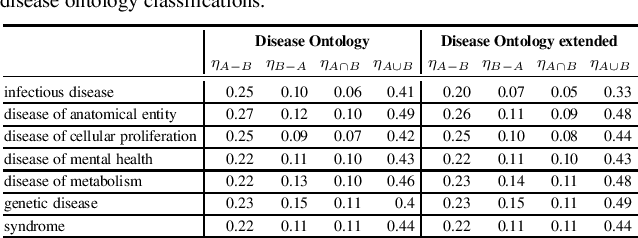
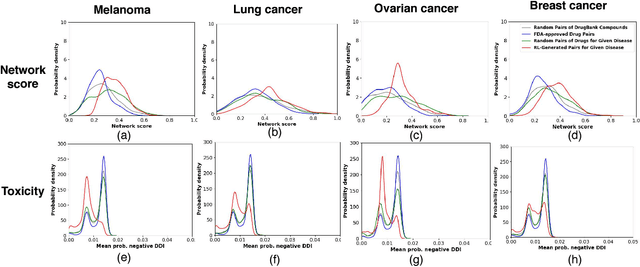
Abstract:Combination therapy has shown to improve therapeutic efficacy while reducing side effects. Importantly, it has become an indispensable strategy to overcome resistance in antibiotics, anti-microbials, and anti-cancer drugs. Facing enormous chemical space and unclear design principles for small-molecule combinations, the computational drug-combination design has not seen generative models to meet its potential to accelerate resistance-overcoming drug combination discovery. We have developed the first deep generative model for drug combination design, by jointly embedding graph-structured domain knowledge and iteratively training a reinforcement learning-based chemical graph-set designer. First, we have developed Hierarchical Variational Graph Auto-Encoders (HVGAE) trained end-to-end to jointly embed gene-gene, gene-disease, and disease-disease networks. Novel attentional pooling is introduced here for learning disease-representations from associated genes' representations. Second, targeting diseases in learned representations, we have recast the drug-combination design problem as graph-set generation and developed a deep learning-based model with novel rewards. Specifically, besides chemical validity rewards, we have introduced a novel generative adversarial award, being generalized sliced Wasserstein, for chemically diverse molecules with distributions similar to known drugs. We have also designed a network principle-based reward for drug combinations. Numerical results indicate that, compared to graph embedding methods, HVGAE learns more informative and generalizable disease representations. Case studies on four diseases show that network-principled drug combinations tend to have low toxicity. The generated drug combinations collectively cover the disease module similar to FDA-approved drug combinations and could potentially suggest novel systems-pharmacology strategies.
Directionally Dependent Multi-View Clustering Using Copula Model
Mar 17, 2020



Abstract:In recent biomedical scientific problems, it is a fundamental issue to integratively cluster a set of objects from multiple sources of datasets. Such problems are mostly encountered in genomics, where data is collected from various sources, and typically represent distinct yet complementary information. Integrating these data sources for multi-source clustering is challenging due to their complex dependence structure including directional dependency. Particularly in genomics studies, it is known that there is certain directional dependence between DNA expression, DNA methylation, and RNA expression, widely called The Central Dogma. Most of the existing multi-view clustering methods either assume an independent structure or pair-wise (non-directional) dependency, thereby ignoring the directional relationship. Motivated by this, we propose a copula-based multi-view clustering model where a copula enables the model to accommodate the directional dependence existing in the datasets. We conduct a simulation experiment where the simulated datasets exhibiting inherent directional dependence: it turns out that ignoring the directional dependence negatively affects the clustering performance. As a real application, we applied our model to the breast cancer tumor samples collected from The Cancer Genome Altas (TCGA).
Explainable Deep Relational Networks for Predicting Compound-Protein Affinities and Contacts
Dec 29, 2019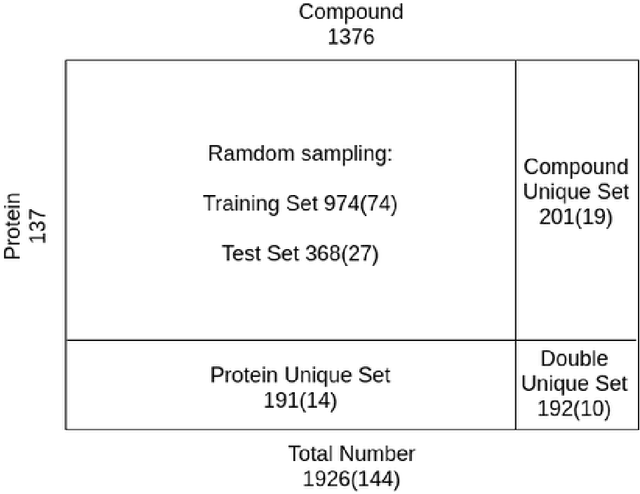
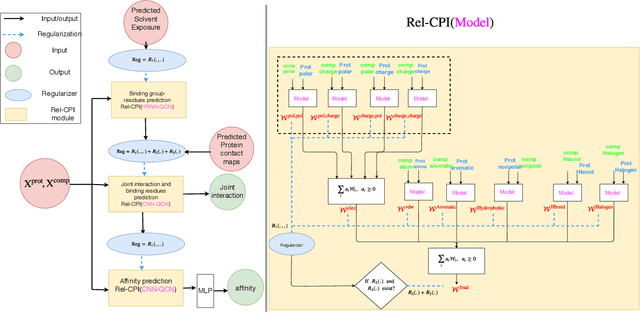
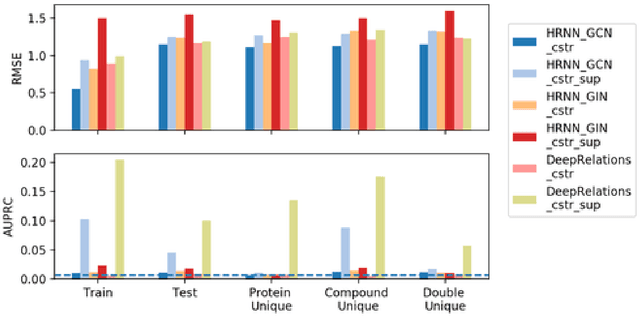
Abstract:Predicting compound-protein affinity is critical for accelerating drug discovery. Recent progress made by machine learning focuses on accuracy but leaves much to be desired for interpretability. Through molecular contacts underlying affinities, our large-scale interpretability assessment finds commonly-used attention mechanisms inadequate. We thus formulate a hierarchical multi-objective learning problem whose predicted contacts form the basis for predicted affinities. We further design a physics-inspired deep relational network, DeepRelations, with intrinsically explainable architecture. Specifically, various atomic-level contacts or "relations" lead to molecular-level affinity prediction. And the embedded attentions are regularized with predicted structural contexts and supervised with partially available training contacts. DeepRelations shows superior interpretability to the state-of-the-art: without compromising affinity prediction, it boosts the AUPRC of contact prediction 9.5, 16.9, 19.3 and 5.7-fold for the test, compound-unique, protein-unique, and both-unique sets, respectively. Our study represents the first dedicated model development and systematic model assessment for interpretable machine learning of compound-protein affinity.
Illegible Text to Readable Text: An Image-to-Image Transformation using Conditional Sliced Wasserstein Adversarial Networks
Oct 11, 2019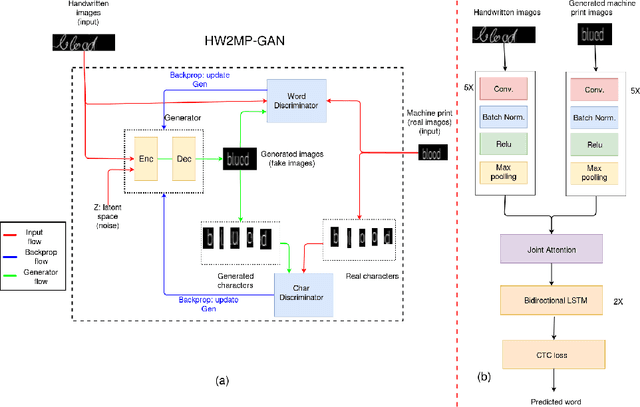
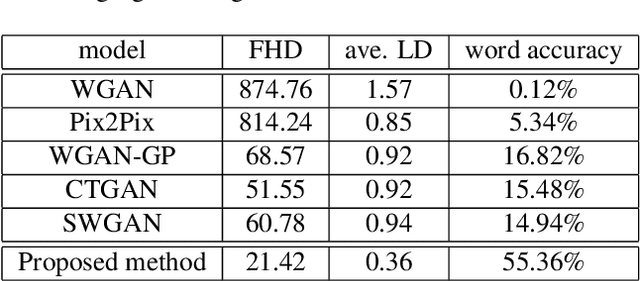


Abstract:Automatic text recognition from ancient handwritten record images is an important problem in the genealogy domain. However, critical challenges such as varying noise conditions, vanishing texts, and variations in handwriting make the recognition task difficult. We tackle this problem by developing a handwritten-to-machine-print conditional Generative Adversarial network (HW2MP-GAN) model that formulates handwritten recognition as a text-Image-to-text-Image translation problem where a given image, typically in an illegible form, is converted into another image, close to its machine-print form. The proposed model consists of three-components including a generator, and word-level and character-level discriminators. The model incorporates Sliced Wasserstein distance (SWD) and U-Net architectures in HW2MP-GAN for better quality image-to-image transformation. Our experiments reveal that HW2MP-GAN outperforms state-of-the-art baseline cGAN models by almost 30 in Frechet Handwritten Distance (FHD), 0.6 on average Levenshtein distance and 39% in word accuracy for image-to-image translation on IAM database. Further, HW2MP-GAN improves handwritten recognition word accuracy by 1.3% compared to baseline handwritten recognition models on the IAM database.
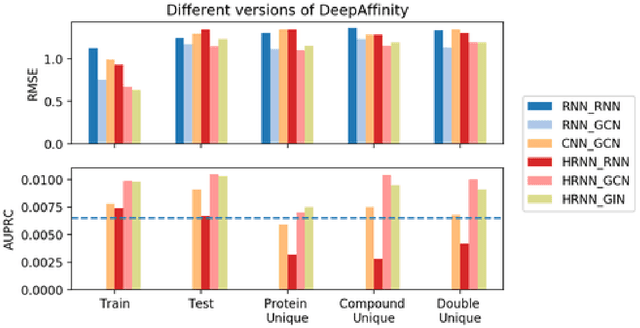
DeepAffinity: Interpretable Deep Learning of Compound-Protein Affinity through Unified Recurrent and Convolutional Neural Networks
Jun 20, 2018

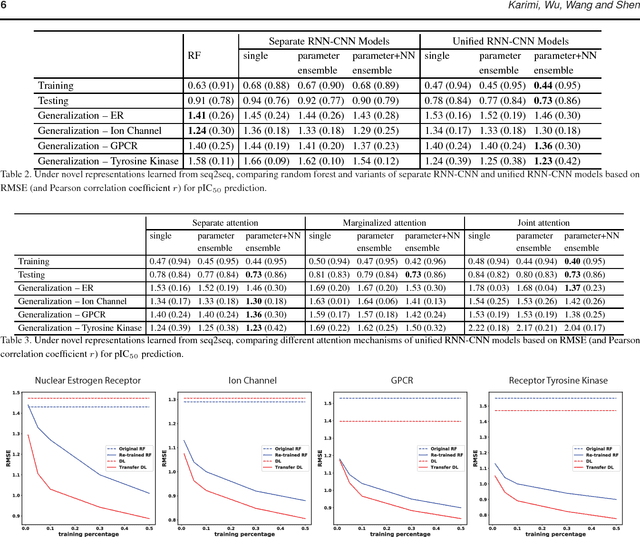
Abstract:Motivation: Drug discovery demands rapid quantification of compound-protein interaction (CPI). However, there is a lack of methods that can predict compound-protein affinity from sequences alone with high applicability, accuracy, and interpretability. Results: We present a seamless integration of domain knowledges and learning-based approaches. Under novel representations of structurally-annotated protein sequences, a semi-supervised deep learning model that unifies recurrent and convolutional neural networks has been proposed to exploit both unlabeled and labeled data, for jointly encoding molecular representations and predicting affinities. Our representations and models outperform conventional options in achieving relative error in IC50 within 5-fold for test cases and 10-fold for protein classes not included for training. Performances for new protein classes with few labeled data are further improved by transfer learning. Furthermore, an attention mechanism is embedded to our model to add to its interpretability, as illustrated in case studies for predicting and explaining selective drug-target interactions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge