DeepAffinity: Interpretable Deep Learning of Compound-Protein Affinity through Unified Recurrent and Convolutional Neural Networks
Paper and Code
Jun 20, 2018

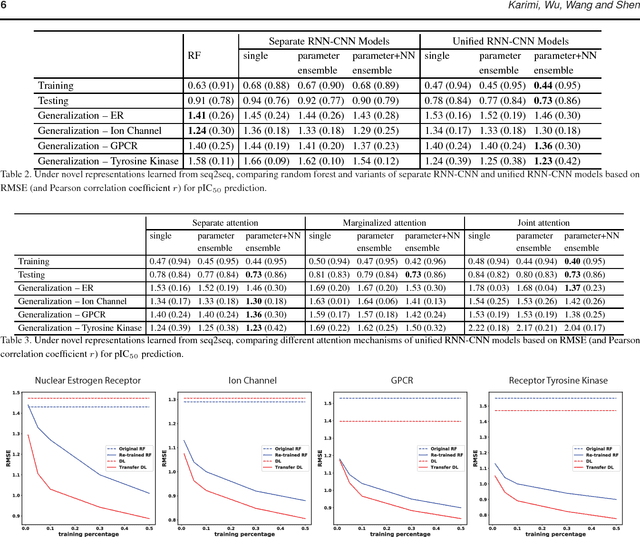
Motivation: Drug discovery demands rapid quantification of compound-protein interaction (CPI). However, there is a lack of methods that can predict compound-protein affinity from sequences alone with high applicability, accuracy, and interpretability. Results: We present a seamless integration of domain knowledges and learning-based approaches. Under novel representations of structurally-annotated protein sequences, a semi-supervised deep learning model that unifies recurrent and convolutional neural networks has been proposed to exploit both unlabeled and labeled data, for jointly encoding molecular representations and predicting affinities. Our representations and models outperform conventional options in achieving relative error in IC50 within 5-fold for test cases and 10-fold for protein classes not included for training. Performances for new protein classes with few labeled data are further improved by transfer learning. Furthermore, an attention mechanism is embedded to our model to add to its interpretability, as illustrated in case studies for predicting and explaining selective drug-target interactions.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge