Se Yoon Lee
Continuous shrinkage prior revisited: a collapsing behavior and remedy
Jul 04, 2020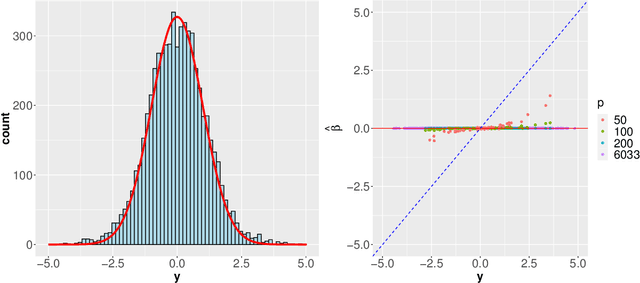
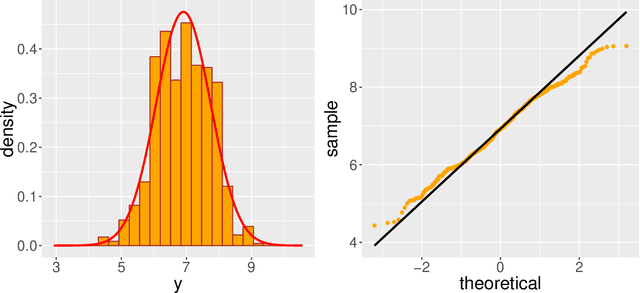
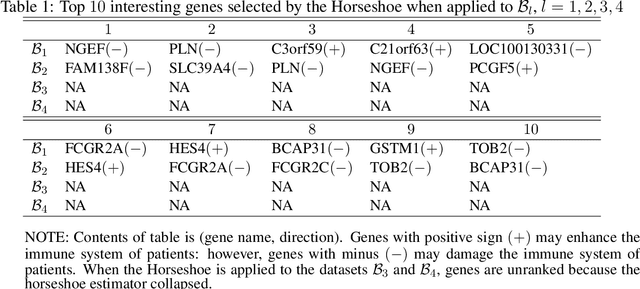
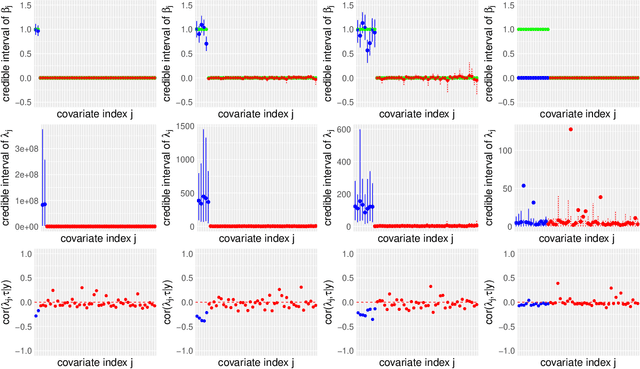
Abstract:Modern genomic studies are increasingly focused on identifying more and more genes clinically associated with a health response. Commonly used Bayesian shrinkage priors are designed primarily to detect only a handful of signals when the dimension of the predictors is very high. In this article, we investigate the performance of a popular continuous shrinkage prior in the presence of relatively large number of true signals. We draw attention to an undesirable phenomenon; the posterior mean is rendered very close to a null vector, caused by a sharp underestimation of the global-scale parameter. The phenomenon is triggered by the absence of a tail-index controlling mechanism in the Bayesian shrinkage priors. We provide a remedy by developing a global-local-tail shrinkage prior which can automatically learn the tail-index and can provide accurate inference even in the presence of moderately large number of signals. The collapsing behavior of the Horseshoe with its remedy is exemplified in numerical examples and in two gene expression datasets.
Directionally Dependent Multi-View Clustering Using Copula Model
Mar 17, 2020



Abstract:In recent biomedical scientific problems, it is a fundamental issue to integratively cluster a set of objects from multiple sources of datasets. Such problems are mostly encountered in genomics, where data is collected from various sources, and typically represent distinct yet complementary information. Integrating these data sources for multi-source clustering is challenging due to their complex dependence structure including directional dependency. Particularly in genomics studies, it is known that there is certain directional dependence between DNA expression, DNA methylation, and RNA expression, widely called The Central Dogma. Most of the existing multi-view clustering methods either assume an independent structure or pair-wise (non-directional) dependency, thereby ignoring the directional relationship. Motivated by this, we propose a copula-based multi-view clustering model where a copula enables the model to accommodate the directional dependence existing in the datasets. We conduct a simulation experiment where the simulated datasets exhibiting inherent directional dependence: it turns out that ignoring the directional dependence negatively affects the clustering performance. As a real application, we applied our model to the breast cancer tumor samples collected from The Cancer Genome Altas (TCGA).
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge