Moritz Kulessa
Combining Predictions under Uncertainty: The Case of Random Decision Trees
Aug 15, 2022Abstract:A common approach to aggregate classification estimates in an ensemble of decision trees is to either use voting or to average the probabilities for each class. The latter takes uncertainty into account, but not the reliability of the uncertainty estimates (so to say, the "uncertainty about the uncertainty"). More generally, much remains unknown about how to best combine probabilistic estimates from multiple sources. In this paper, we investigate a number of alternative prediction methods. Our methods are inspired by the theories of probability, belief functions and reliable classification, as well as a principle that we call evidence accumulation. Our experiments on a variety of data sets are based on random decision trees which guarantees a high diversity in the predictions to be combined. Somewhat unexpectedly, we found that taking the average over the probabilities is actually hard to beat. However, evidence accumulation showed consistently better results on all but very small leafs.
* Preprint version. Appeared in: Discovery Science. 24th International Conference, DS 2021, Halifax, NS, Canada, October 11 to 13, 2021, Proceedings. See https://ds2021.cs.dal.ca/ for further information
Tree-Based Dynamic Classifier Chains
Dec 13, 2021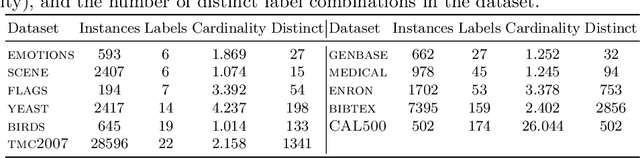
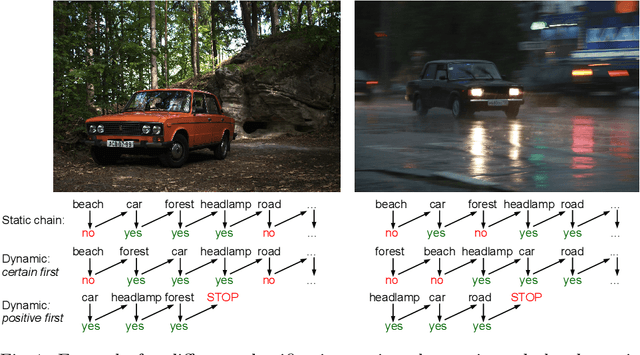
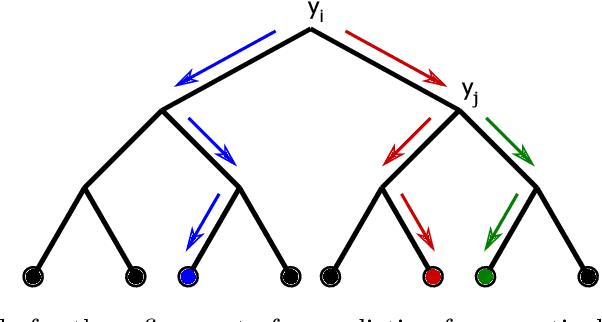
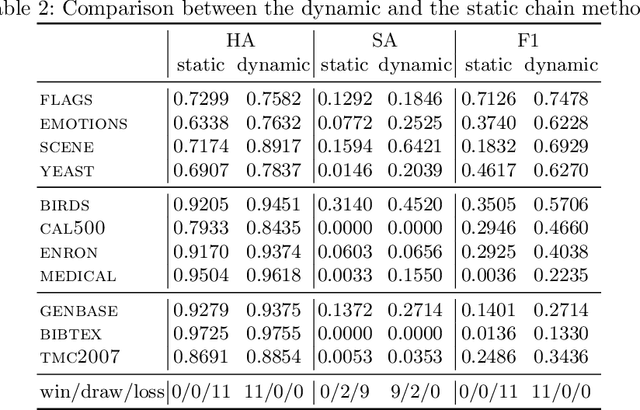
Abstract:Classifier chains are an effective technique for modeling label dependencies in multi-label classification. However, the method requires a fixed, static order of the labels. While in theory, any order is sufficient, in practice, this order has a substantial impact on the quality of the final prediction. Dynamic classifier chains denote the idea that for each instance to classify, the order in which the labels are predicted is dynamically chosen. The complexity of a naive implementation of such an approach is prohibitive, because it would require to train a sequence of classifiers for every possible permutation of the labels. To tackle this problem efficiently, we propose a new approach based on random decision trees which can dynamically select the label ordering for each prediction. We show empirically that a dynamic selection of the next label improves over the use of a static ordering under an otherwise unchanged random decision tree model. % and experimental environment. In addition, we also demonstrate an alternative approach based on extreme gradient boosted trees, which allows for a more target-oriented training of dynamic classifier chains. Our results show that this variant outperforms random decision trees and other tree-based multi-label classification methods. More importantly, the dynamic selection strategy allows to considerably speed up training and prediction.
Correlation-based Discovery of Disease Patterns for Syndromic Surveillance
Oct 18, 2021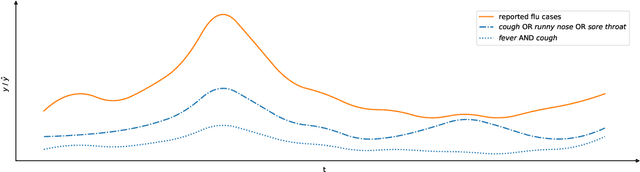
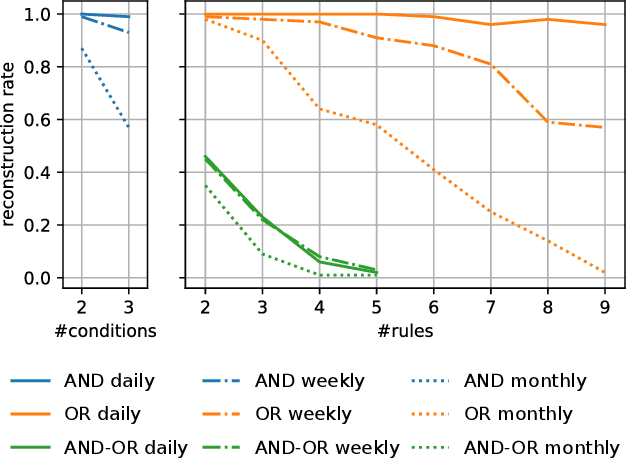
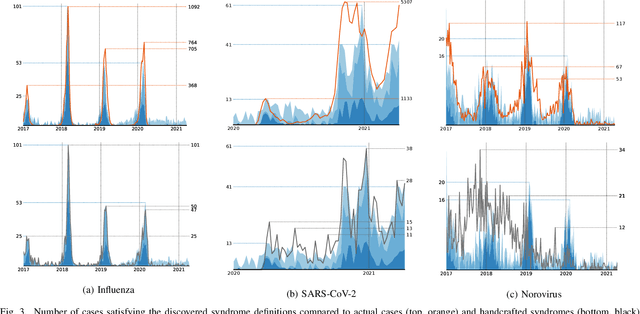
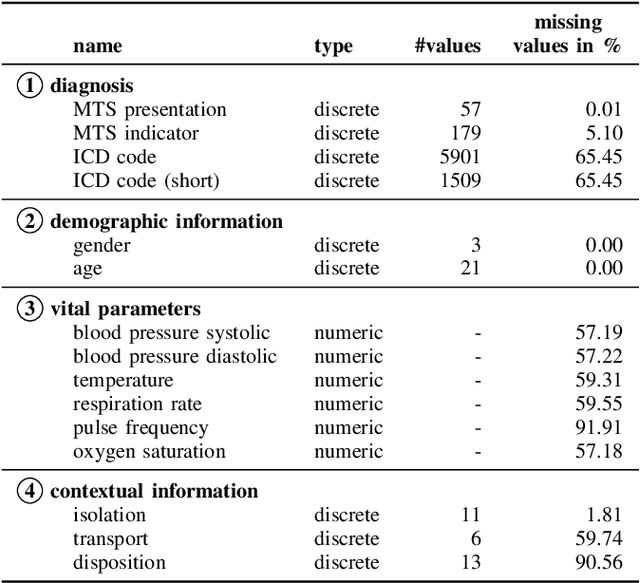
Abstract:Early outbreak detection is a key aspect in the containment of infectious diseases, as it enables the identification and isolation of infected individuals before the disease can spread to a larger population. Instead of detecting unexpected increases of infections by monitoring confirmed cases, syndromic surveillance aims at the detection of cases with early symptoms, which allows a more timely disclosure of outbreaks. However, the definition of these disease patterns is often challenging, as early symptoms are usually shared among many diseases and a particular disease can have several clinical pictures in the early phase of an infection. To support epidemiologists in the process of defining reliable disease patterns, we present a novel, data-driven approach to discover such patterns in historic data. The key idea is to take into account the correlation between indicators in a health-related data source and the reported number of infections in the respective geographic region. In an experimental evaluation, we use data from several emergency departments to discover disease patterns for three infectious diseases. Our results suggest that the proposed approach is able to find patterns that correlate with the reported infections and often identifies indicators that are related to the respective diseases.
Revisiting Non-Specific Syndromic Surveillance
Jan 28, 2021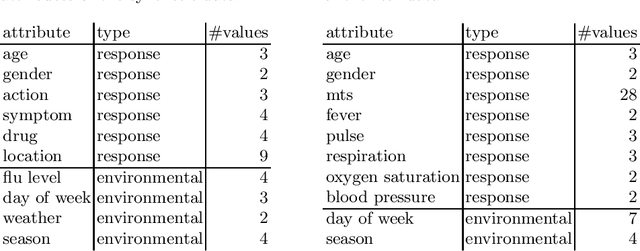

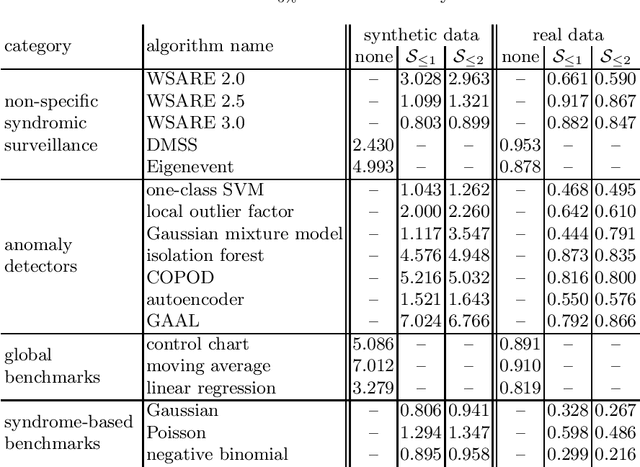
Abstract:Infectious disease surveillance is of great importance for the prevention of major outbreaks. Syndromic surveillance aims at developing algorithms which can detect outbreaks as early as possible by monitoring data sources which allow to capture the occurrences of a certain disease. Recent research mainly focuses on the surveillance of specific, known diseases, putting the focus on the definition of the disease pattern under surveillance. Until now, only little effort has been devoted to what we call non-specific syndromic surveillance, i.e., the use of all available data for detecting any kind of outbreaks, including infectious diseases which are unknown beforehand. In this work, we revisit published approaches for non-specific syndromic surveillance and present a set of simple statistical modeling techniques which can serve as benchmarks for more elaborate machine learning approaches. Our experimental comparison on established synthetic data and real data in which we injected synthetic outbreaks shows that these benchmarks already achieve very competitive results and often outperform more elaborate algorithms.
Improving Outbreak Detection with Stacking of Statistical Surveillance Methods
Jul 17, 2019



Abstract:Epidemiologists use a variety of statistical algorithms for the early detection of outbreaks. The practical usefulness of such methods highly depends on the trade-off between the detection rate of outbreaks and the chances of raising a false alarm. Recent research has shown that the use of machine learning for the fusion of multiple statistical algorithms improves outbreak detection. Instead of relying only on the binary output (alarm or no alarm) of the statistical algorithms, we propose to make use of their p-values for training a fusion classifier. In addition, we also show that adding additional features and adapting the labeling of an epidemic period may further improve performance. For comparison and evaluation, a new measure is introduced which captures the performance of an outbreak detection method with respect to a low rate of false alarms more precisely than previous works. Our results on synthetic data show that it is challenging to improve the performance with a trainable fusion method based on machine learning. In particular, the use of a fusion classifier that is only based on binary outputs of the statistical surveillance methods can make the overall performance worse than directly using the underlying algorithms. However, the use of p-values and additional information for the learning is promising, enabling to identify more valuable patterns to detect outbreaks.
Model-based Approximate Query Processing
Nov 15, 2018



Abstract:Interactive visualizations are arguably the most important tool to explore, understand and convey facts about data. In the past years, the database community has been working on different techniques for Approximate Query Processing (AQP) that aim to deliver an approximate query result given a fixed time bound to support interactive visualizations better. However, classical AQP approaches suffer from various problems that limit the applicability to support the ad-hoc exploration of a new data set: (1) Classical AQP approaches that perform online sampling can support ad-hoc exploration queries but yield low quality if executed over rare subpopulations. (2) Classical AQP approaches that rely on offline sampling can use some form of biased sampling to mitigate these problems but require a priori knowledge of the workload, which is often not realistic if users want to explore a new database. In this paper, we present a new approach to AQP called Model-based AQP that leverages generative models learned over the complete database to answer SQL queries at interactive speeds. Different from classical AQP approaches, generative models allow us to compute responses to ad-hoc queries and deliver high-quality estimates also over rare subpopulations at the same time. In our experiments with real and synthetic data sets, we show that Model-based AQP can in many scenarios return more accurate results in a shorter runtime. Furthermore, we think that our techniques of using generative models presented in this paper can not only be used for AQP in databases but also has applications for other database problems including Query Optimization as well as Data Cleaning.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge