Simon Bohlender
Tree-Based Dynamic Classifier Chains
Dec 13, 2021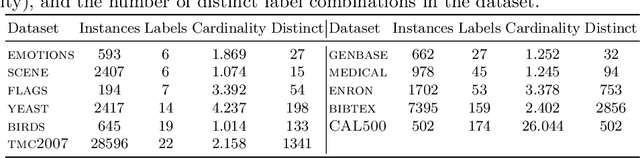
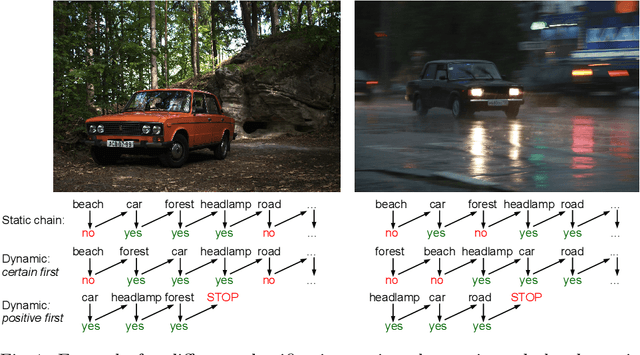
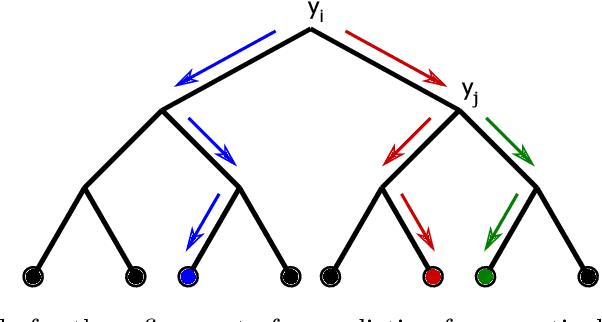
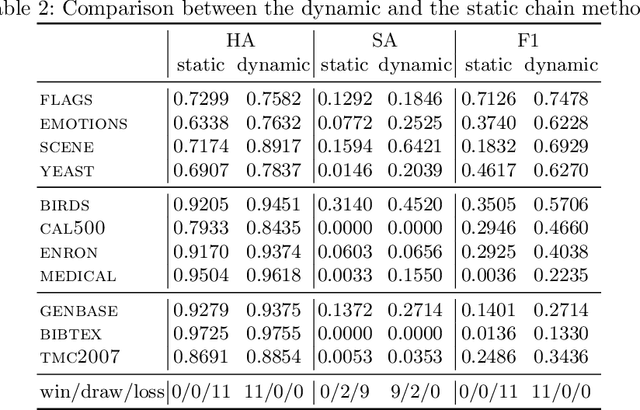
Abstract:Classifier chains are an effective technique for modeling label dependencies in multi-label classification. However, the method requires a fixed, static order of the labels. While in theory, any order is sufficient, in practice, this order has a substantial impact on the quality of the final prediction. Dynamic classifier chains denote the idea that for each instance to classify, the order in which the labels are predicted is dynamically chosen. The complexity of a naive implementation of such an approach is prohibitive, because it would require to train a sequence of classifiers for every possible permutation of the labels. To tackle this problem efficiently, we propose a new approach based on random decision trees which can dynamically select the label ordering for each prediction. We show empirically that a dynamic selection of the next label improves over the use of a static ordering under an otherwise unchanged random decision tree model. % and experimental environment. In addition, we also demonstrate an alternative approach based on extreme gradient boosted trees, which allows for a more target-oriented training of dynamic classifier chains. Our results show that this variant outperforms random decision trees and other tree-based multi-label classification methods. More importantly, the dynamic selection strategy allows to considerably speed up training and prediction.
A survey on shape-constraint deep learning for medical image segmentation
Jan 19, 2021



Abstract:Since the advent of U-Net, fully convolutional deep neural networks and its many variants have completely changed the modern landscape of deep learning based medical image segmentation. However, the over dependence of these methods on pixel level classification and regression has been identified early on as a problem. Especially when trained on medical databases with sparse available annotation, these methods are prone to generate segmentation artifacts such as fragmented structures, topological inconsistencies and islands of pixel. These artefacts are especially problematic in medical imaging since segmentation is almost always a pre-processing step for some downstream evaluation. The range of possible downstream evaluations is rather big, for example surgical planning, visualization, shape analysis, prognosis, treatment planning etc. However, one common thread across all these downstream tasks is the demand of anatomical consistency. To ensure the segmentation result is anatomically consistent, approaches based on Markov/ Conditional Random Fields, Statistical Shape Models are becoming increasingly popular over the past 5 years. In this review paper, a broad overview of recent literature on bringing anatomical constraints for medical image segmentation is given, the shortcomings and opportunities of the proposed methods are thoroughly discussed and potential future work is elaborated. We review the most relevant papers published until the submission date. For quick access, important details such as the underlying method, datasets and performance are tabulated.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge