Revisiting Non-Specific Syndromic Surveillance
Paper and Code
Jan 28, 2021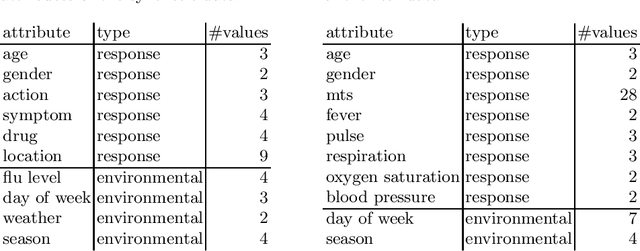

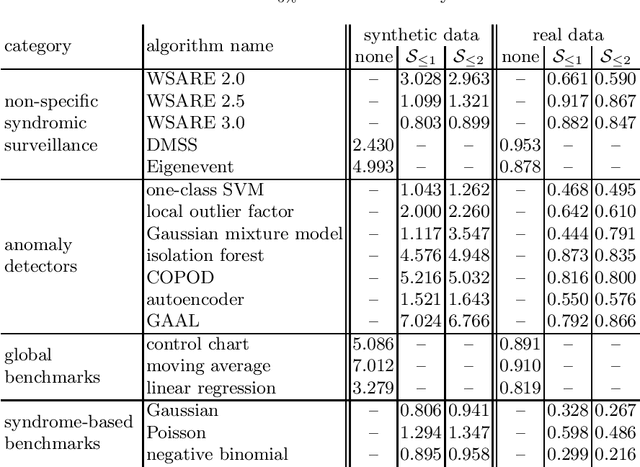
Infectious disease surveillance is of great importance for the prevention of major outbreaks. Syndromic surveillance aims at developing algorithms which can detect outbreaks as early as possible by monitoring data sources which allow to capture the occurrences of a certain disease. Recent research mainly focuses on the surveillance of specific, known diseases, putting the focus on the definition of the disease pattern under surveillance. Until now, only little effort has been devoted to what we call non-specific syndromic surveillance, i.e., the use of all available data for detecting any kind of outbreaks, including infectious diseases which are unknown beforehand. In this work, we revisit published approaches for non-specific syndromic surveillance and present a set of simple statistical modeling techniques which can serve as benchmarks for more elaborate machine learning approaches. Our experimental comparison on established synthetic data and real data in which we injected synthetic outbreaks shows that these benchmarks already achieve very competitive results and often outperform more elaborate algorithms.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge