Mitchell Ostrow
InputDSA: Demixing then Comparing Recurrent and Externally Driven Dynamics
Oct 29, 2025Abstract:In control problems and basic scientific modeling, it is important to compare observations with dynamical simulations. For example, comparing two neural systems can shed light on the nature of emergent computations in the brain and deep neural networks. Recently, Ostrow et al. (2023) introduced Dynamical Similarity Analysis (DSA), a method to measure the similarity of two systems based on their recurrent dynamics rather than geometry or topology. However, DSA does not consider how inputs affect the dynamics, meaning that two similar systems, if driven differently, may be classified as different. Because real-world dynamical systems are rarely autonomous, it is important to account for the effects of input drive. To this end, we introduce a novel metric for comparing both intrinsic (recurrent) and input-driven dynamics, called InputDSA (iDSA). InputDSA extends the DSA framework by estimating and comparing both input and intrinsic dynamic operators using a variant of Dynamic Mode Decomposition with control (DMDc) based on subspace identification. We demonstrate that InputDSA can successfully compare partially observed, input-driven systems from noisy data. We show that when the true inputs are unknown, surrogate inputs can be substituted without a major deterioration in similarity estimates. We apply InputDSA on Recurrent Neural Networks (RNNs) trained with Deep Reinforcement Learning, identifying that high-performing networks are dynamically similar to one another, while low-performing networks are more diverse. Lastly, we apply InputDSA to neural data recorded from rats performing a cognitive task, demonstrating that it identifies a transition from input-driven evidence accumulation to intrinsically-driven decision-making. Our work demonstrates that InputDSA is a robust and efficient method for comparing intrinsic dynamics and the effect of external input on dynamical systems.
Characterizing control between interacting subsystems with deep Jacobian estimation
Jul 02, 2025Abstract:Biological function arises through the dynamical interactions of multiple subsystems, including those between brain areas, within gene regulatory networks, and more. A common approach to understanding these systems is to model the dynamics of each subsystem and characterize communication between them. An alternative approach is through the lens of control theory: how the subsystems control one another. This approach involves inferring the directionality, strength, and contextual modulation of control between subsystems. However, methods for understanding subsystem control are typically linear and cannot adequately describe the rich contextual effects enabled by nonlinear complex systems. To bridge this gap, we devise a data-driven nonlinear control-theoretic framework to characterize subsystem interactions via the Jacobian of the dynamics. We address the challenge of learning Jacobians from time-series data by proposing the JacobianODE, a deep learning method that leverages properties of the Jacobian to directly estimate it for arbitrary dynamical systems from data alone. We show that JacobianODEs outperform existing Jacobian estimation methods on challenging systems, including high-dimensional chaos. Applying our approach to a multi-area recurrent neural network (RNN) trained on a working memory selection task, we show that the "sensory" area gains greater control over the "cognitive" area over learning. Furthermore, we leverage the JacobianODE to directly control the trained RNN, enabling precise manipulation of its behavior. Our work lays the foundation for a theoretically grounded and data-driven understanding of interactions among biological subsystems.
How Diffusion Models Learn to Factorize and Compose
Aug 23, 2024
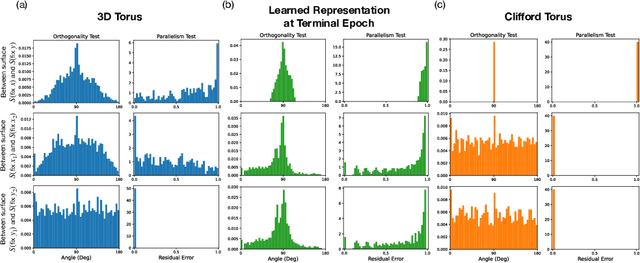

Abstract:Diffusion models are capable of generating photo-realistic images that combine elements which likely do not appear together in the training set, demonstrating the ability to compositionally generalize. Nonetheless, the precise mechanism of compositionality and how it is acquired through training remains elusive. Inspired by cognitive neuroscientific approaches, we consider a highly reduced setting to examine whether and when diffusion models learn semantically meaningful and factorized representations of composable features. We performed extensive controlled experiments on conditional Denoising Diffusion Probabilistic Models (DDPMs) trained to generate various forms of 2D Gaussian data. We found that the models learn factorized but not fully continuous manifold representations for encoding continuous features of variation underlying the data. With such representations, models demonstrate superior feature compositionality but limited ability to interpolate over unseen values of a given feature. Our experimental results further demonstrate that diffusion models can attain compositionality with few compositional examples, suggesting a more efficient way to train DDPMs. Finally, we connect manifold formation in diffusion models to percolation theory in physics, offering insight into the sudden onset of factorized representation learning. Our thorough toy experiments thus contribute a deeper understanding of how diffusion models capture compositional structure in data.
Delay Embedding Theory of Neural Sequence Models
Jun 17, 2024



Abstract:To generate coherent responses, language models infer unobserved meaning from their input text sequence. One potential explanation for this capability arises from theories of delay embeddings in dynamical systems, which prove that unobserved variables can be recovered from the history of only a handful of observed variables. To test whether language models are effectively constructing delay embeddings, we measure the capacities of sequence models to reconstruct unobserved dynamics. We trained 1-layer transformer decoders and state-space sequence models on next-step prediction from noisy, partially-observed time series data. We found that each sequence layer can learn a viable embedding of the underlying system. However, state-space models have a stronger inductive bias than transformers-in particular, they more effectively reconstruct unobserved information at initialization, leading to more parameter-efficient models and lower error on dynamics tasks. Our work thus forges a novel connection between dynamical systems and deep learning sequence models via delay embedding theory.
Bridging Associative Memory and Probabilistic Modeling
Feb 15, 2024Abstract:Associative memory and probabilistic modeling are two fundamental topics in artificial intelligence. The first studies recurrent neural networks designed to denoise, complete and retrieve data, whereas the second studies learning and sampling from probability distributions. Based on the observation that associative memory's energy functions can be seen as probabilistic modeling's negative log likelihoods, we build a bridge between the two that enables useful flow of ideas in both directions. We showcase four examples: First, we propose new energy-based models that flexibly adapt their energy functions to new in-context datasets, an approach we term \textit{in-context learning of energy functions}. Second, we propose two new associative memory models: one that dynamically creates new memories as necessitated by the training data using Bayesian nonparametrics, and another that explicitly computes proportional memory assignments using the evidence lower bound. Third, using tools from associative memory, we analytically and numerically characterize the memory capacity of Gaussian kernel density estimators, a widespread tool in probababilistic modeling. Fourth, we study a widespread implementation choice in transformers -- normalization followed by self attention -- to show it performs clustering on the hypersphere. Altogether, this work urges further exchange of useful ideas between these two continents of artificial intelligence.
Beyond Geometry: Comparing the Temporal Structure of Computation in Neural Circuits with Dynamical Similarity Analysis
Jun 16, 2023Abstract:How can we tell whether two neural networks are utilizing the same internal processes for a particular computation? This question is pertinent for multiple subfields of both neuroscience and machine learning, including neuroAI, mechanistic interpretability, and brain-machine interfaces. Standard approaches for comparing neural networks focus on the spatial geometry of latent states. Yet in recurrent networks, computations are implemented at the level of neural dynamics, which do not have a simple one-to-one mapping with geometry. To bridge this gap, we introduce a novel similarity metric that compares two systems at the level of their dynamics. Our method incorporates two components: Using recent advances in data-driven dynamical systems theory, we learn a high-dimensional linear system that accurately captures core features of the original nonlinear dynamics. Next, we compare these linear approximations via a novel extension of Procrustes Analysis that accounts for how vector fields change under orthogonal transformation. Via four case studies, we demonstrate that our method effectively identifies and distinguishes dynamic structure in recurrent neural networks (RNNs), whereas geometric methods fall short. We additionally show that our method can distinguish learning rules in an unsupervised manner. Our method therefore opens the door to novel data-driven analyses of the temporal structure of neural computation, and to more rigorous testing of RNNs as models of the brain.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge