Ming Guo
DREMnet: An Interpretable Denoising Framework for Semi-Airborne Transient Electromagnetic Signal
Mar 28, 2025
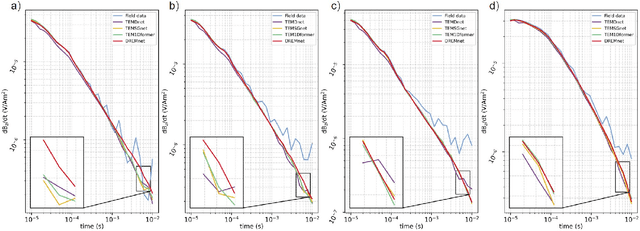


Abstract:The semi-airborne transient electromagnetic method (SATEM) is capable of conducting rapid surveys over large-scale and hard-to-reach areas. However, the acquired signals are often contaminated by complex noise, which can compromise the accuracy of subsequent inversion interpretations. Traditional denoising techniques primarily rely on parameter selection strategies, which are insufficient for processing field data in noisy environments. With the advent of deep learning, various neural networks have been employed for SATEM signal denoising. However, existing deep learning methods typically use single-mapping learning approaches that struggle to effectively separate signal from noise. These methods capture only partial information and lack interpretability. To overcome these limitations, we propose an interpretable decoupled representation learning framework, termed DREMnet, that disentangles data into content and context factors, enabling robust and interpretable denoising in complex conditions. To address the limitations of CNN and Transformer architectures, we utilize the RWKV architecture for data processing and introduce the Contextual-WKV mechanism, which allows unidirectional WKV to perform bidirectional signal modeling. Our proposed Covering Embedding technique retains the strong local perception of convolutional networks through stacked embedding. Experimental results on test datasets demonstrate that the DREMnet method outperforms existing techniques, with processed field data that more accurately reflects the theoretical signal, offering improved identification of subsurface electrical structures.
Multicell-Fold: geometric learning in folding multicellular life
Jul 09, 2024



Abstract:During developmental processes such as embryogenesis, how a group of cells fold into specific structures, is a central question in biology that defines how living organisms form. Establishing tissue-level morphology critically relies on how every single cell decides to position itself relative to its neighboring cells. Despite its importance, it remains a major challenge to understand and predict the behavior of every cell within the living tissue over time during such intricate processes. To tackle this question, we propose a geometric deep learning model that can predict multicellular folding and embryogenesis, accurately capturing the highly convoluted spatial interactions among cells. We demonstrate that multicellular data can be represented with both granular and foam-like physical pictures through a unified graph data structure, considering both cellular interactions and cell junction networks. We successfully use our model to achieve two important tasks, interpretable 4-D morphological sequence alignment, and predicting local cell rearrangements before they occur at single-cell resolution. Furthermore, using an activation map and ablation studies, we demonstrate that cell geometries and cell junction networks together regulate local cell rearrangement which is critical for embryo morphogenesis. This approach provides a novel paradigm to study morphogenesis, highlighting a unified data structure and harnessing the power of geometric deep learning to accurately model the mechanisms and behaviors of cells during development. It offers a pathway toward creating a unified dynamic morphological atlas for a variety of developmental processes such as embryogenesis.
Learning Dynamics from Multicellular Graphs with Deep Neural Networks
Jan 22, 2024Abstract:The inference of multicellular self-assembly is the central quest of understanding morphogenesis, including embryos, organoids, tumors, and many others. However, it has been tremendously difficult to identify structural features that can indicate multicellular dynamics. Here we propose to harness the predictive power of graph-based deep neural networks (GNN) to discover important graph features that can predict dynamics. To demonstrate, we apply a physically informed GNN (piGNN) to predict the motility of multicellular collectives from a snapshot of their positions both in experiments and simulations. We demonstrate that piGNN is capable of navigating through complex graph features of multicellular living systems, which otherwise can not be achieved by classical mechanistic models. With increasing amounts of multicellular data, we propose that collaborative efforts can be made to create a multicellular data bank (MDB) from which it is possible to construct a large multicellular graph model (LMGM) for general-purposed predictions of multicellular organization.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge