Markus J. Buehler
AI-Guided Human-In-the-Loop Inverse Design of High Performance Engineering Structures
Jan 15, 2026Abstract:Inverse design tools such as Topology Optimization (TO) can achieve new levels of improvement for high-performance engineered structures. However, widespread use is hindered by high computational times and a black-box nature that inhibits user interaction. Human-in-the-loop TO approaches are emerging that integrate human intuition into the design generation process. However, these rely on the time-consuming bottleneck of iterative region selection for design modifications. To reduce the number of iterative trials, this contribution presents an AI co-pilot that uses machine learning to predict the user's preferred regions. The prediction model is configured as an image segmentation task with a U-Net architecture. It is trained on synthetic datasets where human preferences either identify the longest topological member or the most complex structural connection. The model successfully predicts plausible regions for modification and presents them to the user as AI recommendations. The human preference model demonstrates generalization across diverse and non-standard TO problems and exhibits emergent behavior outside the single-region selection training data. Demonstration examples show that the new human-in-the-loop TO approach that integrates the AI co-pilot can improve manufacturability or improve the linear buckling load by 39% while only increasing the total design time by 15 sec compared to conventional simplistic TO.
Higher-Order Knowledge Representations for Agentic Scientific Reasoning
Jan 08, 2026Abstract:Scientific inquiry requires systems-level reasoning that integrates heterogeneous experimental data, cross-domain knowledge, and mechanistic evidence into coherent explanations. While Large Language Models (LLMs) offer inferential capabilities, they often depend on retrieval-augmented contexts that lack structural depth. Traditional Knowledge Graphs (KGs) attempt to bridge this gap, yet their pairwise constraints fail to capture the irreducible higher-order interactions that govern emergent physical behavior. To address this, we introduce a methodology for constructing hypergraph-based knowledge representations that faithfully encode multi-entity relationships. Applied to a corpus of ~1,100 manuscripts on biocomposite scaffolds, our framework constructs a global hypergraph of 161,172 nodes and 320,201 hyperedges, revealing a scale-free topology (power law exponent ~1.23) organized around highly connected conceptual hubs. This representation prevents the combinatorial explosion typical of pairwise expansions and explicitly preserves the co-occurrence context of scientific formulations. We further demonstrate that equipping agentic systems with hypergraph traversal tools, specifically using node-intersection constraints, enables them to bridge semantically distant concepts. By exploiting these higher-order pathways, the system successfully generates grounded mechanistic hypotheses for novel composite materials, such as linking cerium oxide to PCL scaffolds via chitosan intermediates. This work establishes a "teacherless" agentic reasoning system where hypergraph topology acts as a verifiable guardrail, accelerating scientific discovery by uncovering relationships obscured by traditional graph methods.
Selective Imperfection as a Generative Framework for Analysis, Creativity and Discovery
Dec 30, 2025Abstract:We introduce materiomusic as a generative framework linking the hierarchical structures of matter with the compositional logic of music. Across proteins, spider webs and flame dynamics, vibrational and architectural principles recur as tonal hierarchies, harmonic progressions, and long-range musical form. Using reversible mappings, from molecular spectra to musical tones and from three-dimensional networks to playable instruments, we show how sound functions as a scientific probe, an epistemic inversion where listening becomes a mode of seeing and musical composition becomes a blueprint for matter. These mappings excavate deep time: patterns originating in femtosecond molecular vibrations or billion-year evolutionary histories become audible. We posit that novelty in science and art emerges when constraints cannot be satisfied within existing degrees of freedom, forcing expansion of the space of viable configurations. Selective imperfection provides the mechanism restoring balance between coherence and adaptability. Quantitative support comes from exhaustive enumeration of all 2^12 musical scales, revealing that culturally significant systems cluster in a mid-entropy, mid-defect corridor, directly paralleling the Hall-Petch optimum where intermediate defect densities maximize material strength. Iterating these mappings creates productive collisions between human creativity and physics, generating new information as musical structures encounter evolutionary constraints. We show how swarm-based AI models compose music exhibiting human-like structural signatures such as small-world connectivity, modular integration, long-range coherence, suggesting a route beyond interpolation toward invention. We show that science and art are generative acts of world-building under constraint, with vibration as a shared grammar organizing structure across scales.
Sparks: Multi-Agent Artificial Intelligence Model Discovers Protein Design Principles
Apr 26, 2025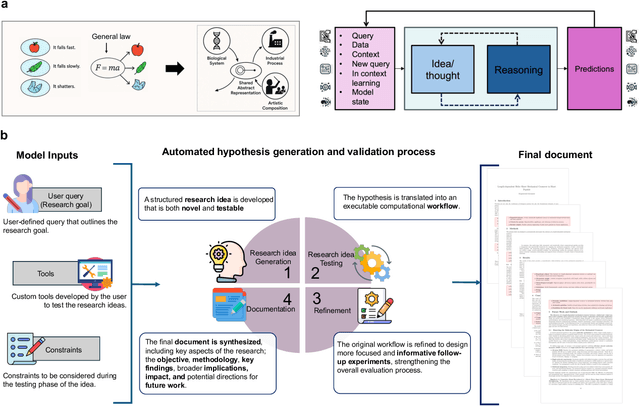
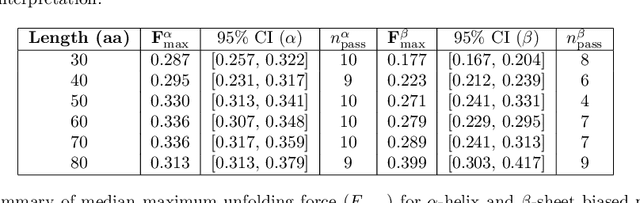

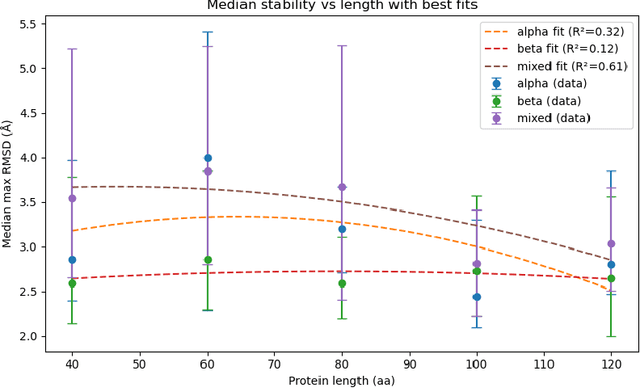
Abstract:Advances in artificial intelligence (AI) promise autonomous discovery, yet most systems still resurface knowledge latent in their training data. We present Sparks, a multi-modal multi-agent AI model that executes the entire discovery cycle that includes hypothesis generation, experiment design and iterative refinement to develop generalizable principles and a report without human intervention. Applied to protein science, Sparks uncovered two previously unknown phenomena: (i) a length-dependent mechanical crossover whereby beta-sheet-biased peptides surpass alpha-helical ones in unfolding force beyond ~80 residues, establishing a new design principle for peptide mechanics; and (ii) a chain-length/secondary-structure stability map revealing unexpectedly robust beta-sheet-rich architectures and a "frustration zone" of high variance in mixed alpha/beta folds. These findings emerged from fully self-directed reasoning cycles that combined generative sequence design, high-accuracy structure prediction and physics-aware property models, with paired generation-and-reflection agents enforcing self-correction and reproducibility. The key result is that Sparks can independently conduct rigorous scientific inquiry and identify previously unknown scientific principles.
Self-Organizing Graph Reasoning Evolves into a Critical State for Continuous Discovery Through Structural-Semantic Dynamics
Mar 24, 2025Abstract:We report fundamental insights into how agentic graph reasoning systems spontaneously evolve toward a critical state that sustains continuous semantic discovery. By rigorously analyzing structural (Von Neumann graph entropy) and semantic (embedding) entropy, we identify a subtle yet robust regime in which semantic entropy persistently dominates over structural entropy. This interplay is quantified by a dimensionless Critical Discovery Parameter that stabilizes at a small negative value, indicating a consistent excess of semantic entropy. Empirically, we observe a stable fraction (12%) of "surprising" edges, links between semantically distant concepts, providing evidence of long-range or cross-domain connections that drive continuous innovation. Concomitantly, the system exhibits scale-free and small-world topological features, alongside a negative cross-correlation between structural and semantic measures, reinforcing the analogy to self-organized criticality. These results establish clear parallels with critical phenomena in physical, biological, and cognitive complex systems, revealing an entropy-based principle governing adaptability and continuous innovation. Crucially, semantic richness emerges as the underlying driver of sustained exploration, despite not being explicitly used by the reasoning process. Our findings provide interdisciplinary insights and practical strategies for engineering intelligent systems with intrinsic capacities for long-term discovery and adaptation, and offer insights into how model training strategies can be developed that reinforce critical discovery.
Agentic Deep Graph Reasoning Yields Self-Organizing Knowledge Networks
Feb 18, 2025



Abstract:We present an agentic, autonomous graph expansion framework that iteratively structures and refines knowledge in situ. Unlike conventional knowledge graph construction methods relying on static extraction or single-pass learning, our approach couples a reasoning-native large language model with a continually updated graph representation. At each step, the system actively generates new concepts and relationships, merges them into a global graph, and formulates subsequent prompts based on its evolving structure. Through this feedback-driven loop, the model organizes information into a scale-free network characterized by hub formation, stable modularity, and bridging nodes that link disparate knowledge clusters. Over hundreds of iterations, new nodes and edges continue to appear without saturating, while centrality measures and shortest path distributions evolve to yield increasingly distributed connectivity. Our analysis reveals emergent patterns, such as the rise of highly connected 'hub' concepts and the shifting influence of 'bridge' nodes, indicating that agentic, self-reinforcing graph construction can yield open-ended, coherent knowledge structures. Applied to materials design problems, we present compositional reasoning experiments by extracting node-specific and synergy-level principles to foster genuinely novel knowledge synthesis, yielding cross-domain ideas that transcend rote summarization and strengthen the framework's potential for open-ended scientific discovery. We discuss other applications in scientific discovery and outline future directions for enhancing scalability and interpretability.
In-situ graph reasoning and knowledge expansion using Graph-PReFLexOR
Jan 14, 2025Abstract:The pursuit of automated scientific discovery has fueled progress from symbolic logic to modern AI, forging new frontiers in reasoning and pattern recognition. Transformers function as potential systems, where every possible relationship remains latent potentiality until tasks impose constraints, akin to measurement. Yet, refining their sampling requires more than probabilistic selection: solutions must conform to specific structures or rules, ensuring consistency and the invocation of general principles. We present Graph-PReFLexOR (Graph-based Preference-based Recursive Language Modeling for Exploratory Optimization of Reasoning), a framework that combines graph reasoning with symbolic abstraction to dynamically expand domain knowledge. Inspired by reinforcement learning, Graph-PReFLexOR defines reasoning as a structured mapping, where tasks yield knowledge graphs, abstract patterns, and ultimately, final answers. Inspired by category theory, it encodes concepts as nodes and their relationships as edges, supporting hierarchical inference and adaptive learning through isomorphic representations. Demonstrations include hypothesis generation, materials design, and creative reasoning, such as discovering relationships between mythological concepts like 'thin places' with materials science. We propose a 'knowledge garden growth' strategy that integrates insights across domains, promoting interdisciplinary connections. Results with a 3-billion-parameter Graph-PReFLexOR model show superior reasoning depth and adaptability, underscoring the potential for transparent, multidisciplinary AI-driven discovery. It lays the groundwork for general autonomous reasoning solutions.
Graph-Aware Isomorphic Attention for Adaptive Dynamics in Transformers
Jan 07, 2025


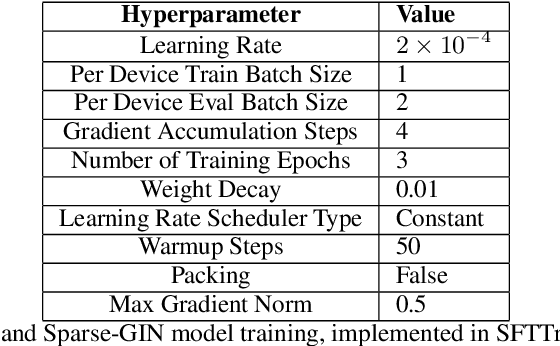
Abstract:We present an approach to modifying Transformer architectures by integrating graph-aware relational reasoning into the attention mechanism, merging concepts from graph neural networks and language modeling. Building on the inherent connection between attention and graph theory, we reformulate the Transformer's attention mechanism as a graph operation and propose Graph-Aware Isomorphic Attention. This method leverages advanced graph modeling strategies, including Graph Isomorphism Networks (GIN) and Principal Neighborhood Aggregation (PNA), to enrich the representation of relational structures. Our approach captures complex dependencies and generalizes across tasks, as evidenced by a reduced generalization gap and improved learning performance. Additionally, we expand the concept of graph-aware attention to introduce Sparse GIN-Attention, a fine-tuning approach that employs sparse GINs. By interpreting attention matrices as sparse adjacency graphs, this technique enhances the adaptability of pre-trained foundational models with minimal computational overhead, endowing them with graph-aware capabilities. Sparse GIN-Attention fine-tuning achieves improved training dynamics and better generalization compared to alternative methods like low-rank adaption (LoRA). We discuss latent graph-like structures within traditional attention mechanisms, offering a new lens through which Transformers can be understood. By evolving Transformers as hierarchical GIN models for relational reasoning. This perspective suggests profound implications for foundational model development, enabling the design of architectures that dynamically adapt to both local and global dependencies. Applications in bioinformatics, materials science, language modeling, and beyond could benefit from this synthesis of relational and sequential data modeling, setting the stage for interpretable and generalizable modeling strategies.
Learning the rules of peptide self-assembly through data mining with large language models
Nov 08, 2024



Abstract:Peptides are ubiquitous and important biologically derived molecules, that have been found to self-assemble to form a wide array of structures. Extensive research has explored the impacts of both internal chemical composition and external environmental stimuli on the self-assembly behaviour of these systems. However, there is yet to be a systematic study that gathers this rich literature data and collectively examines these experimental factors to provide a global picture of the fundamental rules that govern protein self-assembly behavior. In this work, we curate a peptide assembly database through a combination of manual processing by human experts and literature mining facilitated by a large language model. As a result, we collect more than 1,000 experimental data entries with information about peptide sequence, experimental conditions and corresponding self-assembly phases. Utilizing the collected data, ML models are trained and evaluated, demonstrating excellent accuracy (>80\%) and efficiency in peptide assembly phase classification. Moreover, we fine-tune our GPT model for peptide literature mining with the developed dataset, which exhibits markedly superior performance in extracting information from academic publications relative to the pre-trained model. We find that this workflow can substantially improve efficiency when exploring potential self-assembling peptide candidates, through guiding experimental work, while also deepening our understanding of the mechanisms governing peptide self-assembly. In doing so, novel structures can be accessed for a range of applications including sensing, catalysis and biomaterials.
Rapid and Automated Alloy Design with Graph Neural Network-Powered LLM-Driven Multi-Agent Systems
Oct 17, 2024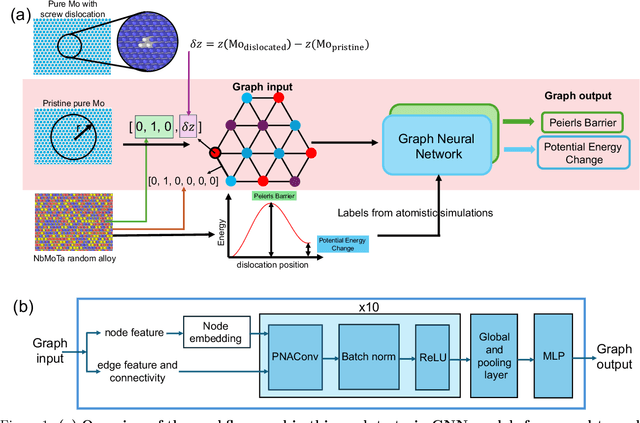
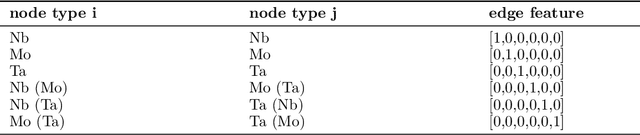
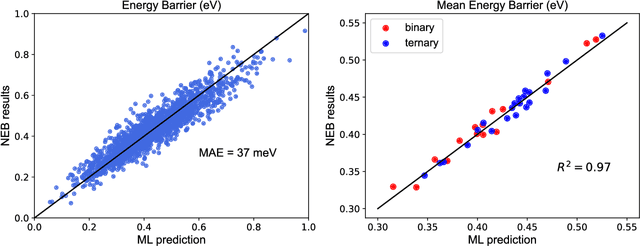
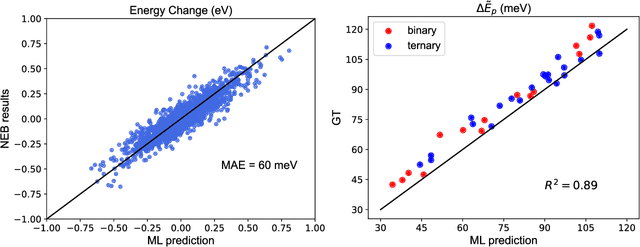
Abstract:A multi-agent AI model is used to automate the discovery of new metallic alloys, integrating multimodal data and external knowledge including insights from physics via atomistic simulations. Our multi-agent system features three key components: (a) a suite of LLMs responsible for tasks such as reasoning and planning, (b) a group of AI agents with distinct roles and expertise that dynamically collaborate, and (c) a newly developed graph neural network (GNN) model for rapid retrieval of key physical properties. A set of LLM-driven AI agents collaborate to automate the exploration of the vast design space of MPEAs, guided by predictions from the GNN. We focus on the NbMoTa family of body-centered cubic (bcc) alloys, modeled using an ML-based interatomic potential, and target two key properties: the Peierls barrier and solute/screw dislocation interaction energy. Our GNN model accurately predicts these atomic-scale properties, providing a faster alternative to costly brute-force calculations and reducing the computational burden on multi-agent systems for physics retrieval. This AI system revolutionizes materials discovery by reducing reliance on human expertise and overcoming the limitations of direct all-atom simulations. By synergizing the predictive power of GNNs with the dynamic collaboration of LLM-based agents, the system autonomously navigates vast alloy design spaces, identifying trends in atomic-scale material properties and predicting macro-scale mechanical strength, as demonstrated by several computational experiments. This approach accelerates the discovery of advanced alloys and holds promise for broader applications in other complex systems, marking a significant step forward in automated materials design.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge