Michael Kim
Enhancing Single-Slice Segmentation with 3D-to-2D Unpaired Scan Distillation
Jun 18, 2024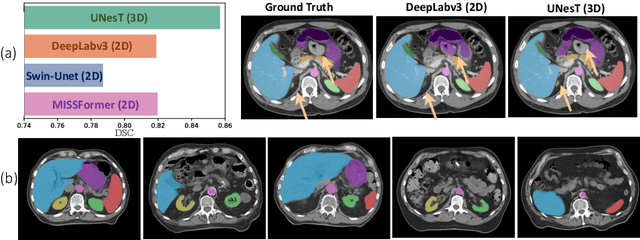
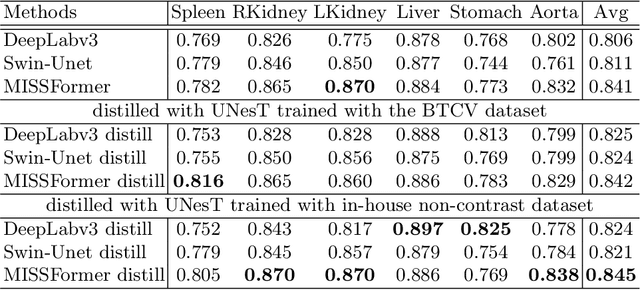
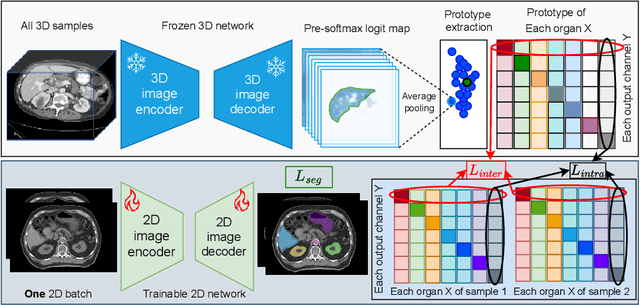
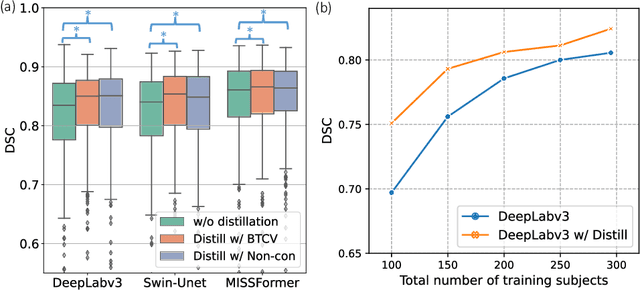
Abstract:2D single-slice abdominal computed tomography (CT) enables the assessment of body habitus and organ health with low radiation exposure. However, single-slice data necessitates the use of 2D networks for segmentation, but these networks often struggle to capture contextual information effectively. Consequently, even when trained on identical datasets, 3D networks typically achieve superior segmentation results. In this work, we propose a novel 3D-to-2D distillation framework, leveraging pre-trained 3D models to enhance 2D single-slice segmentation. Specifically, we extract the prediction distribution centroid from the 3D representations, to guide the 2D student by learning intra- and inter-class correlation. Unlike traditional knowledge distillation methods that require the same data input, our approach employs unpaired 3D CT scans with any contrast to guide the 2D student model. Experiments conducted on 707 subjects from the single-slice Baltimore Longitudinal Study of Aging (BLSA) dataset demonstrate that state-of-the-art 2D multi-organ segmentation methods can benefit from the 3D teacher model, achieving enhanced performance in single-slice multi-organ segmentation. Notably, our approach demonstrates considerable efficacy in low-data regimes, outperforming the model trained with all available training subjects even when utilizing only 200 training subjects. Thus, this work underscores the potential to alleviate manual annotation burdens.
Multi-CATE: Multi-Accurate Conditional Average Treatment Effect Estimation Robust to Unknown Covariate Shifts
May 28, 2024
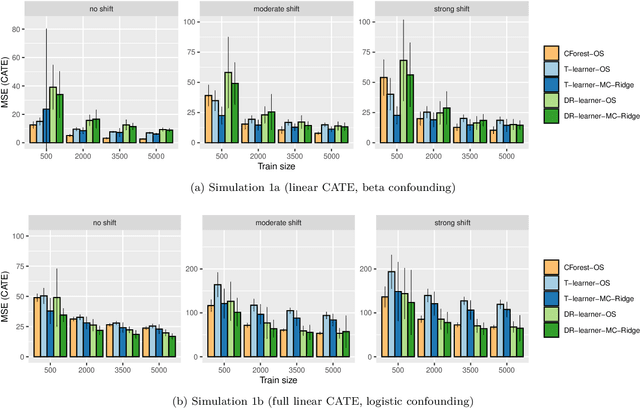
Abstract:Estimating heterogeneous treatment effects is important to tailor treatments to those individuals who would most likely benefit. However, conditional average treatment effect predictors may often be trained on one population but possibly deployed on different, possibly unknown populations. We use methodology for learning multi-accurate predictors to post-process CATE T-learners (differenced regressions) to become robust to unknown covariate shifts at the time of deployment. The method works in general for pseudo-outcome regression, such as the DR-learner. We show how this approach can combine (large) confounded observational and (smaller) randomized datasets by learning a confounded predictor from the observational dataset, and auditing for multi-accuracy on the randomized controlled trial. We show improvements in bias and mean squared error in simulations with increasingly larger covariate shift, and on a semi-synthetic case study of a parallel large observational study and smaller randomized controlled experiment. Overall, we establish a connection between methods developed for multi-distribution learning and achieve appealing desiderata (e.g. external validity) in causal inference and machine learning.
Field-of-View Extension for Diffusion MRI via Deep Generative Models
May 06, 2024



Abstract:Purpose: In diffusion MRI (dMRI), the volumetric and bundle analyses of whole-brain tissue microstructure and connectivity can be severely impeded by an incomplete field-of-view (FOV). This work aims to develop a method for imputing the missing slices directly from existing dMRI scans with an incomplete FOV. We hypothesize that the imputed image with complete FOV can improve the whole-brain tractography for corrupted data with incomplete FOV. Therefore, our approach provides a desirable alternative to discarding the valuable dMRI data, enabling subsequent tractography analyses that would otherwise be challenging or unattainable with corrupted data. Approach: We propose a framework based on a deep generative model that estimates the absent brain regions in dMRI scans with incomplete FOV. The model is capable of learning both the diffusion characteristics in diffusion-weighted images (DWI) and the anatomical features evident in the corresponding structural images for efficiently imputing missing slices of DWI outside of incomplete FOV. Results: For evaluating the imputed slices, on the WRAP dataset the proposed framework achieved PSNRb0=22.397, SSIMb0=0.905, PSNRb1300=22.479, SSIMb1300=0.893; on the NACC dataset it achieved PSNRb0=21.304, SSIMb0=0.892, PSNRb1300=21.599, SSIMb1300= 0.877. The proposed framework improved the tractography accuracy, as demonstrated by an increased average Dice score for 72 tracts (p < 0.001) on both the WRAP and NACC datasets. Conclusions: Results suggest that the proposed framework achieved sufficient imputation performance in dMRI data with incomplete FOV for improving whole-brain tractography, thereby repairing the corrupted data. Our approach achieved more accurate whole-brain tractography results with extended and complete FOV and reduced the uncertainty when analyzing bundles associated with Alzheimer's Disease.
Generalized Linear Tree Space Nearest Neighbor
Mar 30, 2021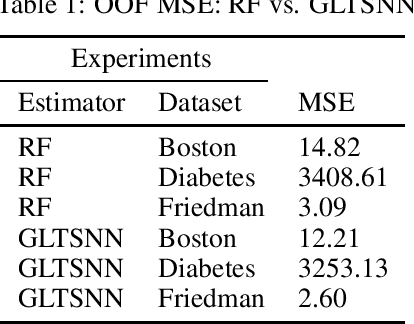
Abstract:We present a novel method of stacking decision trees by projection into an ordered time split out-of-fold (OOF) one nearest neighbor (1NN) space. The predictions of these one nearest neighbors are combined through a linear model. This process is repeated many times and averaged to reduce variance. Generalized Linear Tree Space Nearest Neighbor (GLTSNN) is competitive with respect to Mean Squared Error (MSE) compared to Random Forest (RF) on several publicly available datasets. Some of the theoretical and applied advantages of GLTSNN are discussed. We conjecture a classifier based upon the GLTSNN would have an error that is asymptotically bounded by twice the Bayes error rate like k = 1 Nearest Neighbor.
Developing Quantum Annealer Driven Data Discovery
Mar 25, 2016
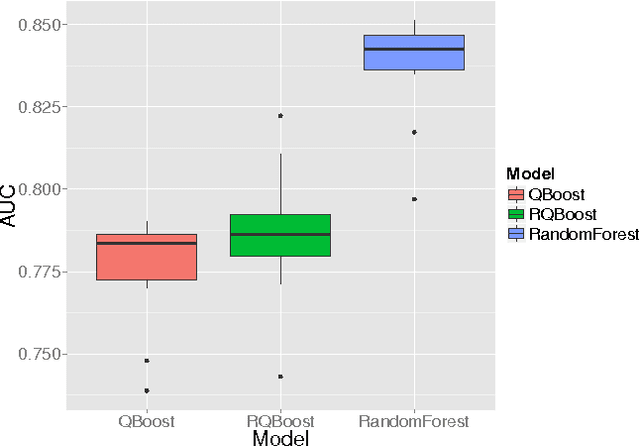
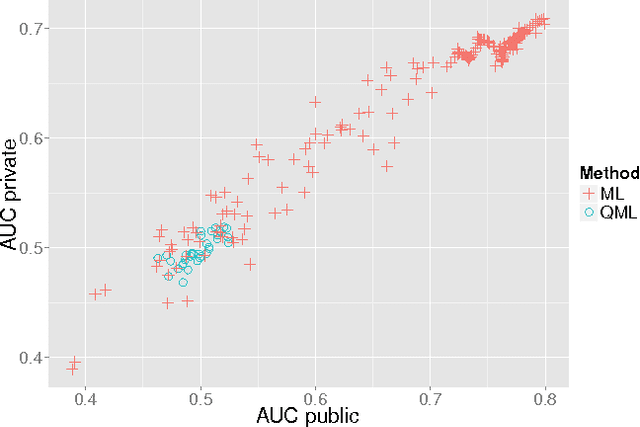
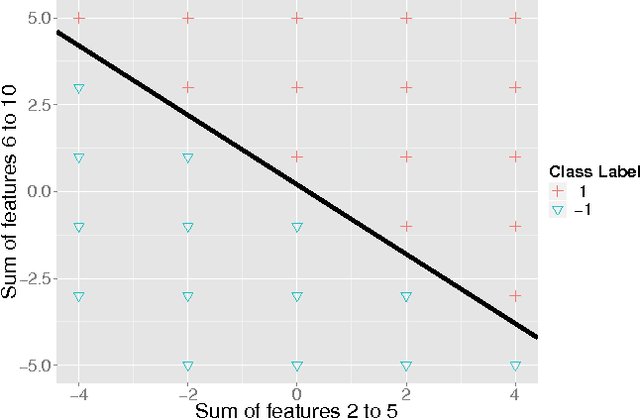
Abstract:Machine learning applications are limited by computational power. In this paper, we gain novel insights into the application of quantum annealing (QA) to machine learning (ML) through experiments in natural language processing (NLP), seizure prediction, and linear separability testing. These experiments are performed on QA simulators and early-stage commercial QA hardware and compared to an unprecedented number of traditional ML techniques. We extend QBoost, an early implementation of a binary classifier that utilizes a quantum annealer, via resampling and ensembling of predicted probabilities to produce a more robust class estimator. To determine the strengths and weaknesses of this approach, resampled QBoost (RQBoost) is tested across several datasets and compared to QBoost and traditional ML. We show and explain how QBoost in combination with a commercial QA device are unable to perfectly separate binary class data which is linearly separable via logistic regression with shrinkage. We further explore the performance of RQBoost in the space of NLP and seizure prediction and find QA-enabled ML using QBoost and RQBoost is outperformed by traditional techniques. Additionally, we provide a detailed discussion of algorithmic constraints and trade-offs imposed by the use of this QA hardware. Through these experiments, we provide unique insights into the state of quantum ML via boosting and the use of quantum annealing hardware that are valuable to institutions interested in applying QA to problems in ML and beyond.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge