Matthew Ricci
MODE: Learning compositional representations of complex systems with Mixtures Of Dynamical Experts
Oct 10, 2025



Abstract:Dynamical systems in the life sciences are often composed of complex mixtures of overlapping behavioral regimes. Cellular subpopulations may shift from cycling to equilibrium dynamics or branch towards different developmental fates. The transitions between these regimes can appear noisy and irregular, posing a serious challenge to traditional, flow-based modeling techniques which assume locally smooth dynamics. To address this challenge, we propose MODE (Mixture Of Dynamical Experts), a graphical modeling framework whose neural gating mechanism decomposes complex dynamics into sparse, interpretable components, enabling both the unsupervised discovery of behavioral regimes and accurate long-term forecasting across regime transitions. Crucially, because agents in our framework can jump to different governing laws, MODE is especially tailored to the aforementioned noisy transitions. We evaluate our method on a battery of synthetic and real datasets from computational biology. First, we systematically benchmark MODE on an unsupervised classification task using synthetic dynamical snapshot data, including in noisy, few-sample settings. Next, we show how MODE succeeds on challenging forecasting tasks which simulate key cycling and branching processes in cell biology. Finally, we deploy our method on human, single-cell RNA sequencing data and show that it can not only distinguish proliferation from differentiation dynamics but also predict when cells will commit to their ultimate fate, a key outstanding challenge in computational biology.
Characterizing Nonlinear Dynamics via Smooth Prototype Equivalences
Mar 13, 2025



Abstract:Characterizing dynamical systems given limited measurements is a common challenge throughout the physical and biological sciences. However, this task is challenging, especially due to transient variability in systems with equivalent long-term dynamics. We address this by introducing smooth prototype equivalences (SPE), a framework that fits a diffeomorphism using normalizing flows to distinct prototypes - simplified dynamical systems that define equivalence classes of behavior. SPE enables classification by comparing the deformation loss of the observed sparse, high-dimensional measurements to the prototype dynamics. Furthermore, our approach enables estimation of the invariant sets of the observed dynamics through the learned mapping from prototype space to data space. Our method outperforms existing techniques in the classification of oscillatory systems and can efficiently identify invariant structures like limit cycles and fixed points in an equation-free manner, even when only a small, noisy subset of the phase space is observed. Finally, we show how our method can be used for the detection of biological processes like the cell cycle trajectory from high-dimensional single-cell gene expression data.
TRENDy: Temporal Regression of Effective Non-linear Dynamics
Dec 04, 2024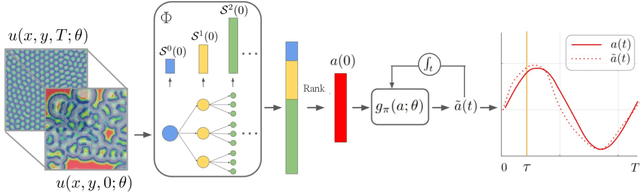
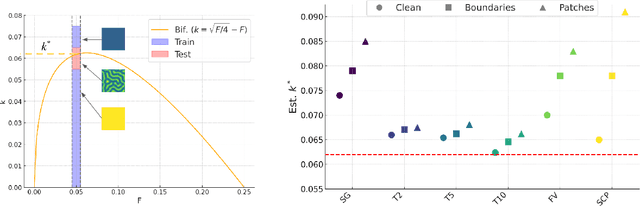
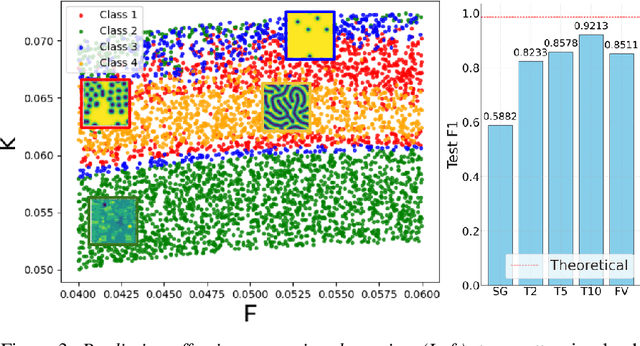
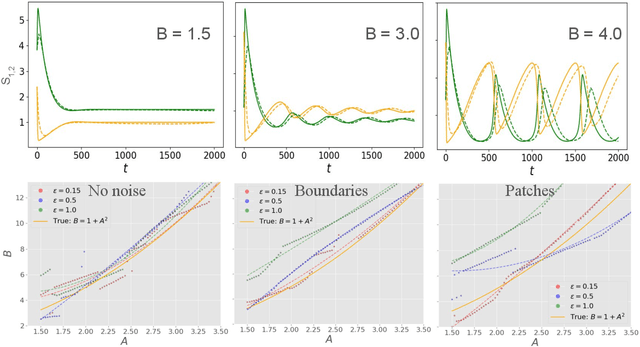
Abstract:Spatiotemporal dynamics pervade the natural sciences, from the morphogen dynamics underlying patterning in animal pigmentation to the protein waves controlling cell division. A central challenge lies in understanding how controllable parameters induce qualitative changes in system behavior called bifurcations. This endeavor is made particularly difficult in realistic settings where governing partial differential equations (PDEs) are unknown and data is limited and noisy. To address this challenge, we propose TRENDy (Temporal Regression of Effective Nonlinear Dynamics), an equation-free approach to learning low-dimensional, predictive models of spatiotemporal dynamics. Following classical work in spatial coarse-graining, TRENDy first maps input data to a low-dimensional space of effective dynamics via a cascade of multiscale filtering operations. Our key insight is the recognition that these effective dynamics can be fit by a neural ordinary differential equation (NODE) having the same parameter space as the input PDE. The preceding filtering operations strongly regularize the phase space of the NODE, making TRENDy significantly more robust to noise compared to existing methods. We train TRENDy to predict the effective dynamics of synthetic and real data representing dynamics from across the physical and life sciences. We then demonstrate how our framework can automatically locate both Turing and Hopf bifurcations in unseen regions of parameter space. We finally apply our method to the analysis of spatial patterning of the ocellated lizard through development. We found that TRENDy's effective state not only accurately predicts spatial changes over time but also identifies distinct pattern features unique to different anatomical regions, highlighting the potential influence of surface geometry on reaction-diffusion mechanisms and their role in driving spatially varying pattern dynamics.
Let's do the time-warp-attend: Learning topological invariants of dynamical systems
Dec 14, 2023



Abstract:Dynamical systems across the sciences, from electrical circuits to ecological networks, undergo qualitative and often catastrophic changes in behavior, called bifurcations, when their underlying parameters cross a threshold. Existing methods predict oncoming catastrophes in individual systems but are primarily time-series-based and struggle both to categorize qualitative dynamical regimes across diverse systems and to generalize to real data. To address this challenge, we propose a data-driven, physically-informed deep-learning framework for classifying dynamical regimes and characterizing bifurcation boundaries based on the extraction of topologically invariant features. We focus on the paradigmatic case of the supercritical Hopf bifurcation, which is used to model periodic dynamics across a wide range of applications. Our convolutional attention method is trained with data augmentations that encourage the learning of topological invariants which can be used to detect bifurcation boundaries in unseen systems and to design models of biological systems like oscillatory gene regulatory networks. We further demonstrate our method's use in analyzing real data by recovering distinct proliferation and differentiation dynamics along pancreatic endocrinogenesis trajectory in gene expression space based on single-cell data. Our method provides valuable insights into the qualitative, long-term behavior of a wide range of dynamical systems, and can detect bifurcations or catastrophic transitions in large-scale physical and biological systems.
Phase2vec: Dynamical systems embedding with a physics-informed convolutional network
Dec 07, 2022



Abstract:Dynamical systems are found in innumerable forms across the physical and biological sciences, yet all these systems fall naturally into universal equivalence classes: conservative or dissipative, stable or unstable, compressible or incompressible. Predicting these classes from data remains an essential open challenge in computational physics at which existing time-series classification methods struggle. Here, we propose, \texttt{phase2vec}, an embedding method that learns high-quality, physically-meaningful representations of 2D dynamical systems without supervision. Our embeddings are produced by a convolutional backbone that extracts geometric features from flow data and minimizes a physically-informed vector field reconstruction loss. In an auxiliary training period, embeddings are optimized so that they robustly encode the equations of unseen data over and above the performance of a per-equation fitting method. The trained architecture can not only predict the equations of unseen data, but also, crucially, learns embeddings that respect the underlying semantics of the embedded physical systems. We validate the quality of learned embeddings investigating the extent to which physical categories of input data can be decoded from embeddings compared to standard blackbox classifiers and state-of-the-art time series classification techniques. We find that our embeddings encode important physical properties of the underlying data, including the stability of fixed points, conservation of energy, and the incompressibility of flows, with greater fidelity than competing methods. We finally apply our embeddings to the analysis of meteorological data, showing we can detect climatically meaningful features. Collectively, our results demonstrate the viability of embedding approaches for the discovery of dynamical features in physical systems.
KuraNet: Systems of Coupled Oscillators that Learn to Synchronize
May 06, 2021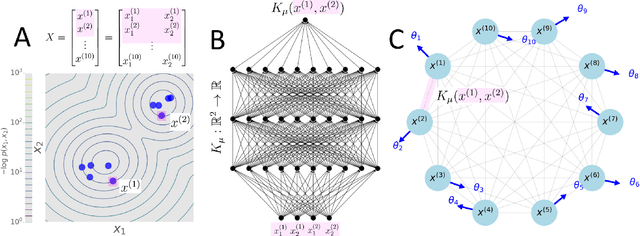
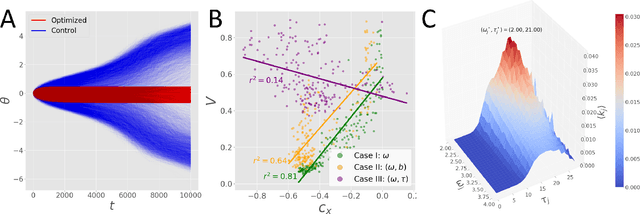
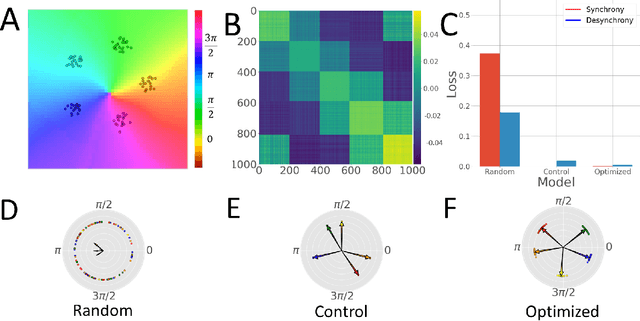
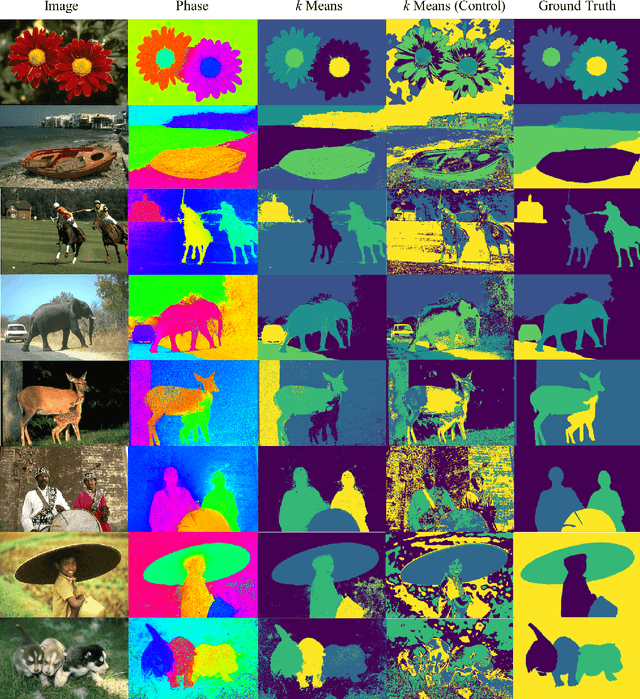
Abstract:Networks of coupled oscillators are some of the most studied objects in the theory of dynamical systems. Two important areas of current interest are the study of synchrony in highly disordered systems and the modeling of systems with adaptive network structures. Here, we present a single approach to both of these problems in the form of "KuraNet", a deep-learning-based system of coupled oscillators that can learn to synchronize across a distribution of disordered network conditions. The key feature of the model is the replacement of the traditionally static couplings with a coupling function which can learn optimal interactions within heterogeneous oscillator populations. We apply our approach to the eponymous Kuramoto model and demonstrate how KuraNet can learn data-dependent coupling structures that promote either global or cluster synchrony. For example, we show how KuraNet can be used to empirically explore the conditions of global synchrony in analytically impenetrable models with disordered natural frequencies, external field strengths, and interaction delays. In a sequence of cluster synchrony experiments, we further show how KuraNet can function as a data classifier by synchronizing into coherent assemblies. In all cases, we show how KuraNet can generalize to both new data and new network scales, making it easy to work with small systems and form hypotheses about the thermodynamic limit. Our proposed learning-based approach is broadly applicable to arbitrary dynamical systems with wide-ranging relevance to modeling in physics and systems biology.
Neural Optimal Control for Representation Learning
Jun 16, 2020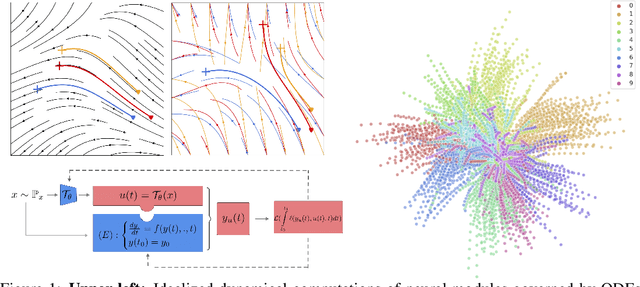
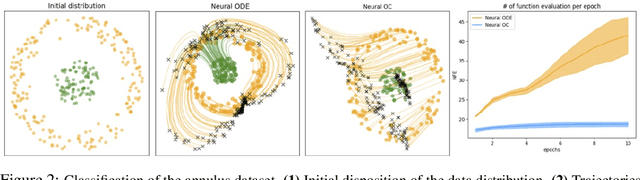


Abstract:The intriguing connections recently established between neural networks and dynamical systems have invited deep learning researchers to tap into the well-explored principles of differential calculus. Notably, the adjoint sensitivity method used in neural ordinary differential equations (Neural ODEs) has cast the training of neural networks as a control problem in which neural modules operate as continuous-time homeomorphic transformations of features. Typically, these methods optimize a single set of parameters governing the dynamical system for the whole data set, forcing the network to learn complex transformations that are functionally limited and computationally heavy. Instead, we propose learning a data-conditioned distribution of \emph{optimal controls} over the network dynamics, emulating a form of input-dependent fast neural plasticity. We describe a general method for training such models as well as convergence proofs assuming mild hypotheses about the ODEs and show empirically that this method leads to simpler dynamics and reduces the computational cost of Neural ODEs. We evaluate this approach for unsupervised image representation learning; our new "functional" auto-encoding model with ODEs, AutoencODE, achieves state-of-the-art image reconstruction quality on CIFAR-10, and exhibits substantial improvements in unsupervised classification over existing auto-encoding models.
Same-different problems strain convolutional neural networks
May 25, 2018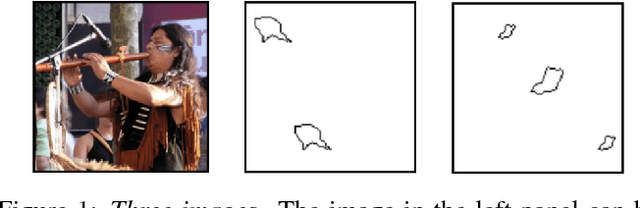
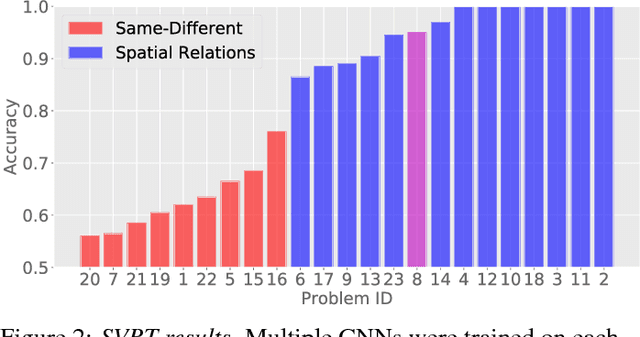
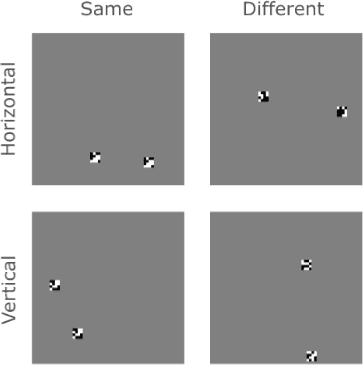
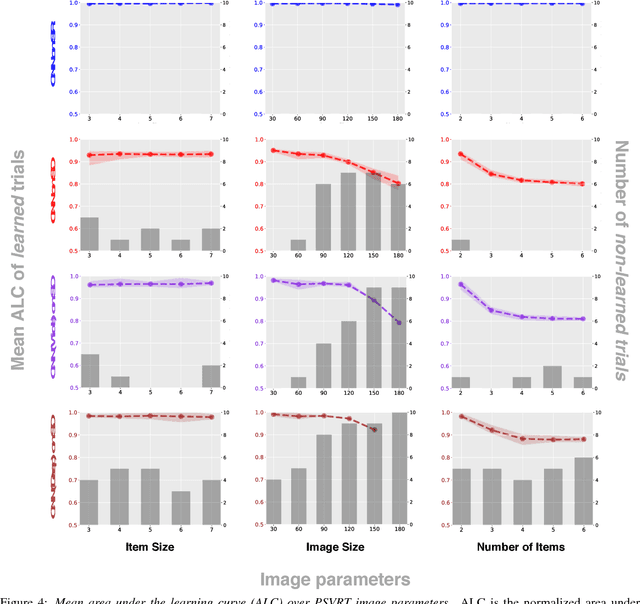
Abstract:The robust and efficient recognition of visual relations in images is a hallmark of biological vision. We argue that, despite recent progress in visual recognition, modern machine vision algorithms are severely limited in their ability to learn visual relations. Through controlled experiments, we demonstrate that visual-relation problems strain convolutional neural networks (CNNs). The networks eventually break altogether when rote memorization becomes impossible, as when intra-class variability exceeds network capacity. Motivated by the comparable success of biological vision, we argue that feedback mechanisms including attention and perceptual grouping may be the key computational components underlying abstract visual reasoning.\
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge