Guy Pelc
Characterizing Nonlinear Dynamics via Smooth Prototype Equivalences
Mar 13, 2025



Abstract:Characterizing dynamical systems given limited measurements is a common challenge throughout the physical and biological sciences. However, this task is challenging, especially due to transient variability in systems with equivalent long-term dynamics. We address this by introducing smooth prototype equivalences (SPE), a framework that fits a diffeomorphism using normalizing flows to distinct prototypes - simplified dynamical systems that define equivalence classes of behavior. SPE enables classification by comparing the deformation loss of the observed sparse, high-dimensional measurements to the prototype dynamics. Furthermore, our approach enables estimation of the invariant sets of the observed dynamics through the learned mapping from prototype space to data space. Our method outperforms existing techniques in the classification of oscillatory systems and can efficiently identify invariant structures like limit cycles and fixed points in an equation-free manner, even when only a small, noisy subset of the phase space is observed. Finally, we show how our method can be used for the detection of biological processes like the cell cycle trajectory from high-dimensional single-cell gene expression data.
TRENDy: Temporal Regression of Effective Non-linear Dynamics
Dec 04, 2024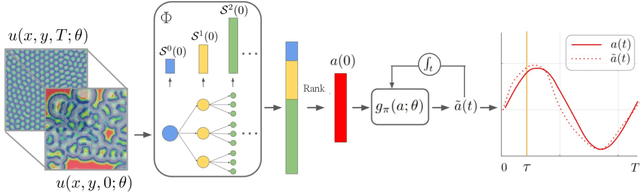
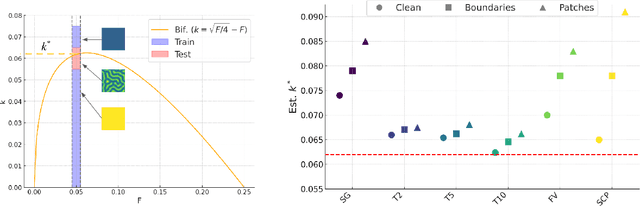
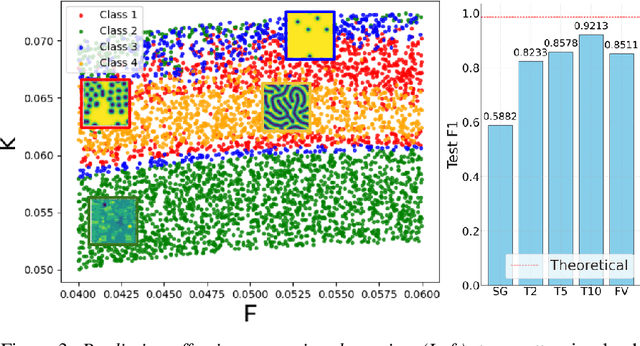
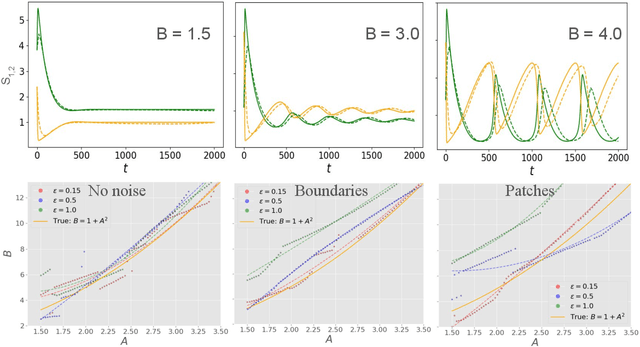
Abstract:Spatiotemporal dynamics pervade the natural sciences, from the morphogen dynamics underlying patterning in animal pigmentation to the protein waves controlling cell division. A central challenge lies in understanding how controllable parameters induce qualitative changes in system behavior called bifurcations. This endeavor is made particularly difficult in realistic settings where governing partial differential equations (PDEs) are unknown and data is limited and noisy. To address this challenge, we propose TRENDy (Temporal Regression of Effective Nonlinear Dynamics), an equation-free approach to learning low-dimensional, predictive models of spatiotemporal dynamics. Following classical work in spatial coarse-graining, TRENDy first maps input data to a low-dimensional space of effective dynamics via a cascade of multiscale filtering operations. Our key insight is the recognition that these effective dynamics can be fit by a neural ordinary differential equation (NODE) having the same parameter space as the input PDE. The preceding filtering operations strongly regularize the phase space of the NODE, making TRENDy significantly more robust to noise compared to existing methods. We train TRENDy to predict the effective dynamics of synthetic and real data representing dynamics from across the physical and life sciences. We then demonstrate how our framework can automatically locate both Turing and Hopf bifurcations in unseen regions of parameter space. We finally apply our method to the analysis of spatial patterning of the ocellated lizard through development. We found that TRENDy's effective state not only accurately predicts spatial changes over time but also identifies distinct pattern features unique to different anatomical regions, highlighting the potential influence of surface geometry on reaction-diffusion mechanisms and their role in driving spatially varying pattern dynamics.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge