Matias Nicolas Bossa
The STOIC2021 COVID-19 AI challenge: applying reusable training methodologies to private data
Jun 25, 2023Abstract:Challenges drive the state-of-the-art of automated medical image analysis. The quantity of public training data that they provide can limit the performance of their solutions. Public access to the training methodology for these solutions remains absent. This study implements the Type Three (T3) challenge format, which allows for training solutions on private data and guarantees reusable training methodologies. With T3, challenge organizers train a codebase provided by the participants on sequestered training data. T3 was implemented in the STOIC2021 challenge, with the goal of predicting from a computed tomography (CT) scan whether subjects had a severe COVID-19 infection, defined as intubation or death within one month. STOIC2021 consisted of a Qualification phase, where participants developed challenge solutions using 2000 publicly available CT scans, and a Final phase, where participants submitted their training methodologies with which solutions were trained on CT scans of 9724 subjects. The organizers successfully trained six of the eight Final phase submissions. The submitted codebases for training and running inference were released publicly. The winning solution obtained an area under the receiver operating characteristic curve for discerning between severe and non-severe COVID-19 of 0.815. The Final phase solutions of all finalists improved upon their Qualification phase solutions.HSUXJM-TNZF9CHSUXJM-TNZF9C
On the Composition and Limitations of Publicly Available COVID-19 X-Ray Imaging Datasets
Aug 26, 2020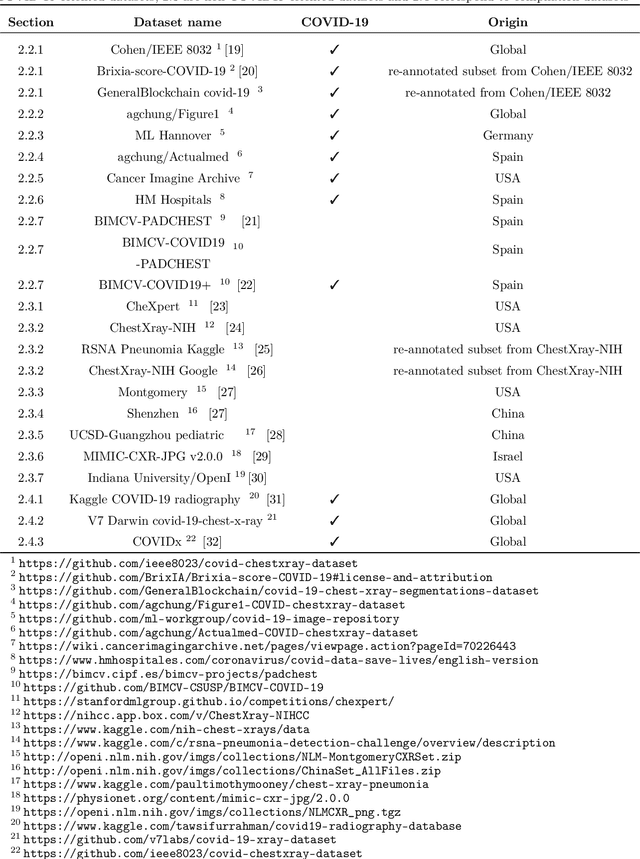
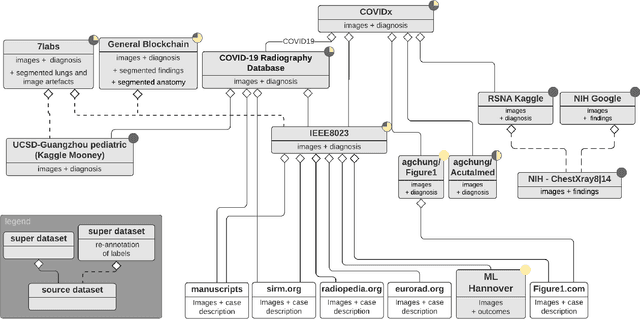
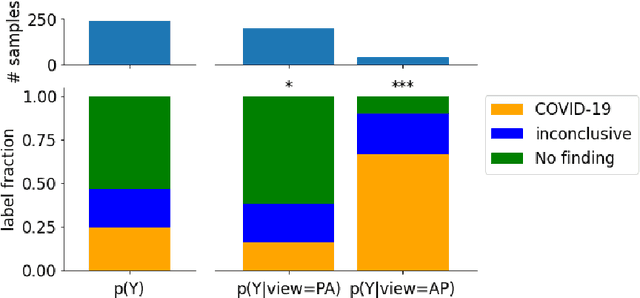
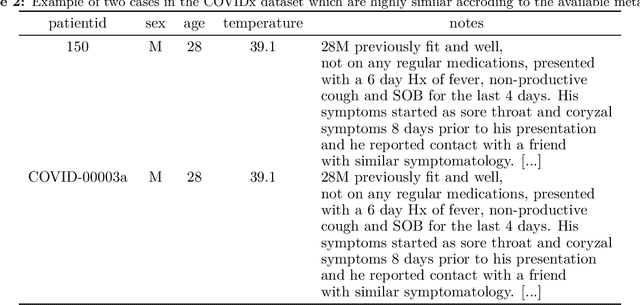
Abstract:Machine learning based methods for diagnosis and progression prediction of COVID-19 from imaging data have gained significant attention in the last months, in particular by the use of deep learning models. In this context hundreds of models where proposed with the majority of them trained on public datasets. Data scarcity, mismatch between training and target population, group imbalance, and lack of documentation are important sources of bias, hindering the applicability of these models to real-world clinical practice. Considering that datasets are an essential part of model building and evaluation, a deeper understanding of the current landscape is needed. This paper presents an overview of the currently public available COVID-19 chest X-ray datasets. Each dataset is briefly described and potential strength, limitations and interactions between datasets are identified. In particular, some key properties of current datasets that could be potential sources of bias, impairing models trained on them are pointed out. These descriptions are useful for model building on those datasets, to choose the best dataset according the model goal, to take into account the specific limitations to avoid reporting overconfident benchmark results, and to discuss their impact on the generalisation capabilities in a specific clinical setting
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge