Mary A. Rutherford
An automated pipeline for quantitative T2* fetal body MRI and segmentation at low field
Aug 09, 2023Abstract:Fetal Magnetic Resonance Imaging at low field strengths is emerging as an exciting direction in perinatal health. Clinical low field (0.55T) scanners are beneficial for fetal imaging due to their reduced susceptibility-induced artefacts, increased T2* values, and wider bore (widening access for the increasingly obese pregnant population). However, the lack of standard automated image processing tools such as segmentation and reconstruction hampers wider clinical use. In this study, we introduce a semi-automatic pipeline using quantitative MRI for the fetal body at low field strength resulting in fast and detailed quantitative T2* relaxometry analysis of all major fetal body organs. Multi-echo dynamic sequences of the fetal body were acquired and reconstructed into a single high-resolution volume using deformable slice-to-volume reconstruction, generating both structural and quantitative T2* 3D volumes. A neural network trained using a semi-supervised approach was created to automatically segment these fetal body 3D volumes into ten different organs (resulting in dice values > 0.74 for 8 out of 10 organs). The T2* values revealed a strong relationship with GA in the lungs, liver, and kidney parenchyma (R^2>0.5). This pipeline was used successfully for a wide range of GAs (17-40 weeks), and is robust to motion artefacts. Low field fetal MRI can be used to perform advanced MRI analysis, and is a viable option for clinical scanning.
Self-supervised Recurrent Neural Network for 4D Abdominal and In-utero MR Imaging
Aug 28, 2019



Abstract:Accurately estimating and correcting the motion artifacts are crucial for 3D image reconstruction of the abdominal and in-utero magnetic resonance imaging (MRI). The state-of-art methods are based on slice-to-volume registration (SVR) where multiple 2D image stacks are acquired in three orthogonal orientations. In this work, we present a novel reconstruction pipeline that only needs one orientation of 2D MRI scans and can reconstruct the full high-resolution image without masking or registration steps. The framework consists of two main stages: the respiratory motion estimation using a self-supervised recurrent neural network, which learns the respiratory signals that are naturally embedded in the asymmetry relationship of the neighborhood slices and cluster them according to a respiratory state. Then, we train a 3D deconvolutional network for super-resolution (SR) reconstruction of the sparsely selected 2D images using integrated reconstruction and total variation loss. We evaluate the classification accuracy on 5 simulated images and compare our results with the SVR method in adult abdominal and in-utero MRI scans. The results show that the proposed pipeline can accurately estimate the respiratory state and reconstruct 4D SR volumes with better or similar performance to the 3D SVR pipeline with less than 20\% sparsely selected slices. The method has great potential to transform the 4D abdominal and in-utero MRI in clinical practice.
DeepCut: Object Segmentation from Bounding Box Annotations using Convolutional Neural Networks
Jun 05, 2016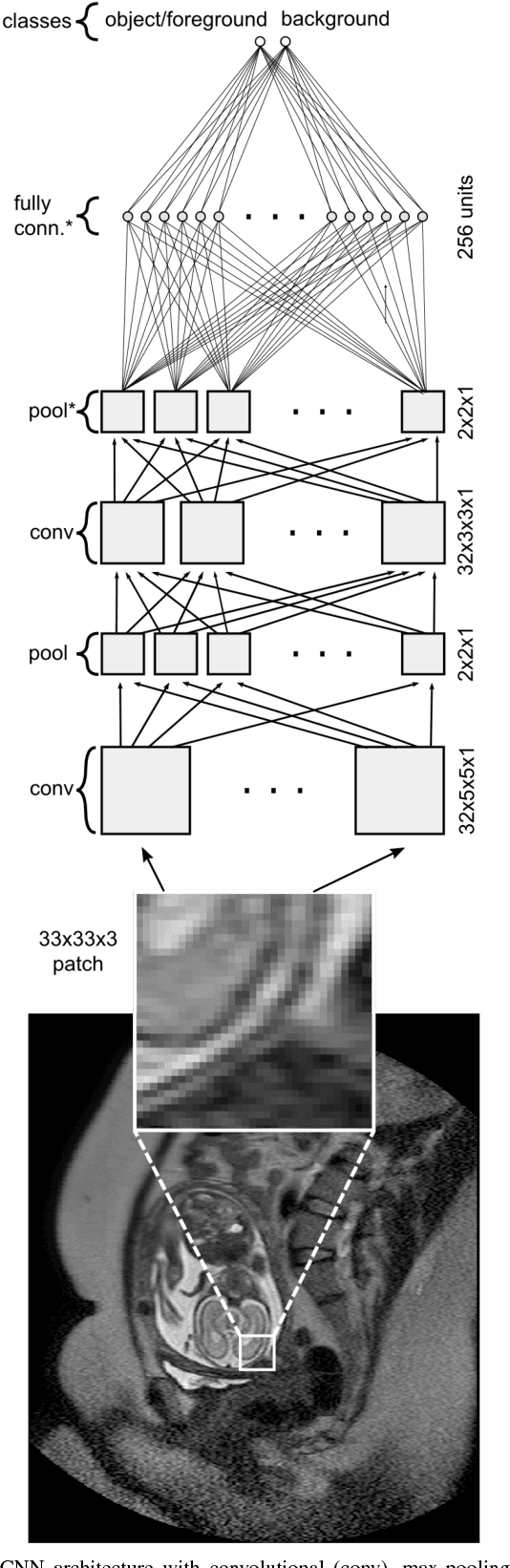
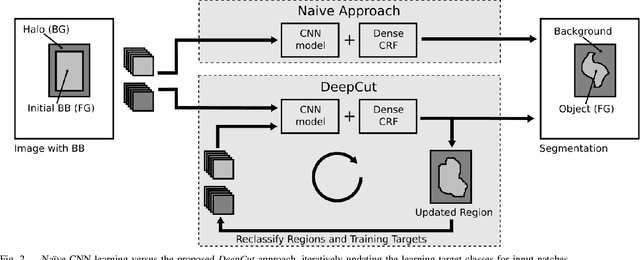
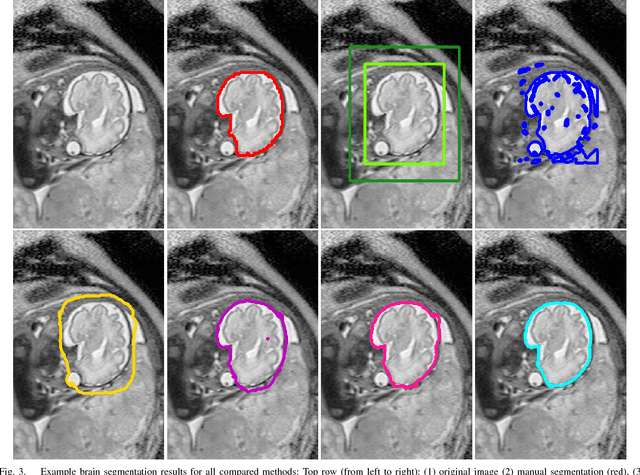
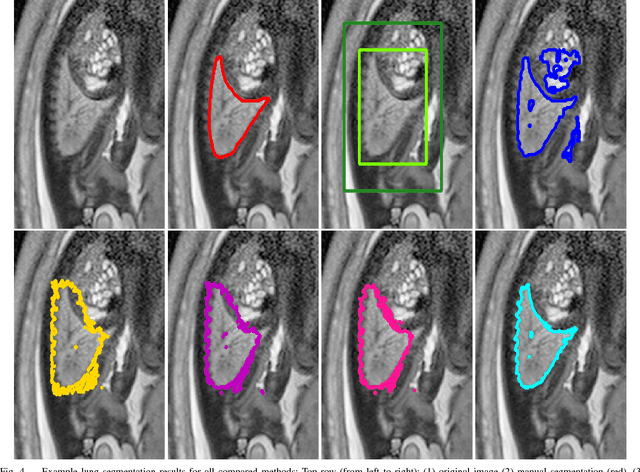
Abstract:In this paper, we propose DeepCut, a method to obtain pixelwise object segmentations given an image dataset labelled with bounding box annotations. It extends the approach of the well-known GrabCut method to include machine learning by training a neural network classifier from bounding box annotations. We formulate the problem as an energy minimisation problem over a densely-connected conditional random field and iteratively update the training targets to obtain pixelwise object segmentations. Additionally, we propose variants of the DeepCut method and compare those to a naive approach to CNN training under weak supervision. We test its applicability to solve brain and lung segmentation problems on a challenging fetal magnetic resonance dataset and obtain encouraging results in terms of accuracy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge