Marine Le Morvan
SODA
TabICLv2: A better, faster, scalable, and open tabular foundation model
Feb 11, 2026Abstract:Tabular foundation models, such as TabPFNv2 and TabICL, have recently dethroned gradient-boosted trees at the top of predictive benchmarks, demonstrating the value of in-context learning for tabular data. We introduce TabICLv2, a new state-of-the-art foundation model for regression and classification built on three pillars: (1) a novel synthetic data generation engine designed for high pretraining diversity; (2) various architectural innovations, including a new scalable softmax in attention improving generalization to larger datasets without prohibitive long-sequence pretraining; and (3) optimized pretraining protocols, notably replacing AdamW with the Muon optimizer. On the TabArena and TALENT benchmarks, TabICLv2 without any tuning surpasses the performance of the current state of the art, RealTabPFN-2.5 (hyperparameter-tuned, ensembled, and fine-tuned on real data). With only moderate pretraining compute, TabICLv2 generalizes effectively to million-scale datasets under 50GB GPU memory while being markedly faster than RealTabPFN-2.5. We provide extensive ablation studies to quantify these contributions and commit to open research by first releasing inference code and model weights at https://github.com/soda-inria/tabicl, with synthetic data engine and pretraining code to follow.
TabICL: A Tabular Foundation Model for In-Context Learning on Large Data
Feb 08, 2025



Abstract:The long-standing dominance of gradient-boosted decision trees on tabular data is currently challenged by tabular foundation models using In-Context Learning (ICL): setting the training data as context for the test data and predicting in a single forward pass without parameter updates. While the very recent TabPFNv2 foundation model (2025) excels on tables with up to 10K samples, its alternating column- and row-wise attentions make handling large training sets computationally prohibitive. So, can ICL be effectively scaled and deliver a benefit for larger tables? We introduce TabICL, a tabular foundation model for classification, pretrained on synthetic datasets with up to 60K samples and capable of handling 500K samples on affordable resources. This is enabled by a novel two-stage architecture: a column-then-row attention mechanism to build fixed-dimensional embeddings of rows, followed by a transformer for efficient ICL. Across 200 classification datasets from the TALENT benchmark, TabICL is on par with TabPFNv2 while being systematically faster (up to 10 times), and significantly outperforms all other approaches. On 56 datasets with over 10K samples, TabICL surpasses both TabPFNv2 and CatBoost, demonstrating the potential of ICL for large data.
Imputation for prediction: beware of diminishing returns
Jul 29, 2024



Abstract:Missing values are prevalent across various fields, posing challenges for training and deploying predictive models. In this context, imputation is a common practice, driven by the hope that accurate imputations will enhance predictions. However, recent theoretical and empirical studies indicate that simple constant imputation can be consistent and competitive. This empirical study aims at clarifying if and when investing in advanced imputation methods yields significantly better predictions. Relating imputation and predictive accuracies across combinations of imputation and predictive models on 20 datasets, we show that imputation accuracy matters less i) when using expressive models, ii) when incorporating missingness indicators as complementary inputs, iii) matters much more for generated linear outcomes than for real-data outcomes. Interestingly, we also show that the use of the missingness indicator is beneficial to the prediction performance, even in MCAR scenarios. Overall, on real-data with powerful models, improving imputation only has a minor effect on prediction performance. Thus, investing in better imputations for improved predictions often offers limited benefits.
Beyond calibration: estimating the grouping loss of modern neural networks
Oct 28, 2022Abstract:Good decision making requires machine-learning models to provide trustworthy confidence scores. To this end, recent work has focused on miscalibration, i.e, the over or under confidence of model scores. Yet, contrary to widespread belief, calibration is not enough: even a classifier with the best possible accuracy and perfect calibration can have confidence scores far from the true posterior probabilities. This is due to the grouping loss, created by samples with the same confidence scores but different true posterior probabilities. Proper scoring rule theory shows that given the calibration loss, the missing piece to characterize individual errors is the grouping loss. While there are many estimators of the calibration loss, none exists for the grouping loss in standard settings. Here, we propose an estimator to approximate the grouping loss. We use it to study modern neural network architectures in vision and NLP. We find that the grouping loss varies markedly across architectures, and that it is a key model-comparison factor across the most accurate, calibrated, models. We also show that distribution shifts lead to high grouping loss.
Benchmarking missing-values approaches for predictive models on health databases
Feb 17, 2022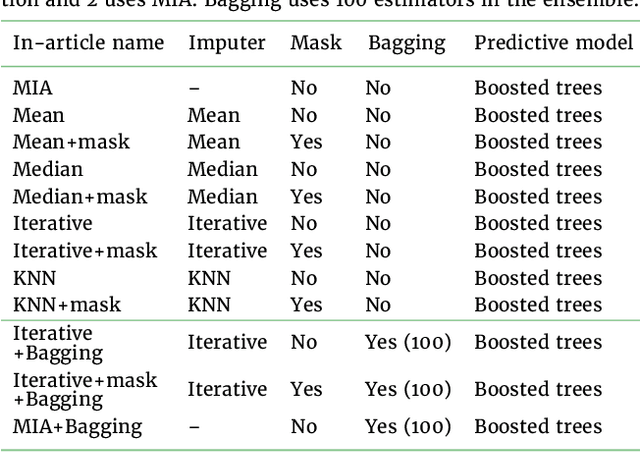
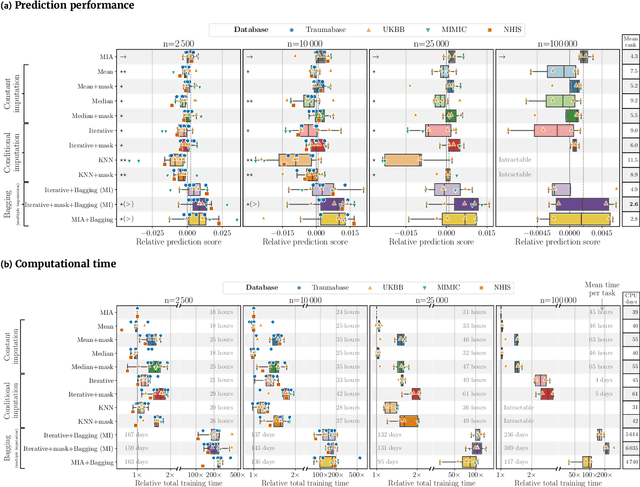
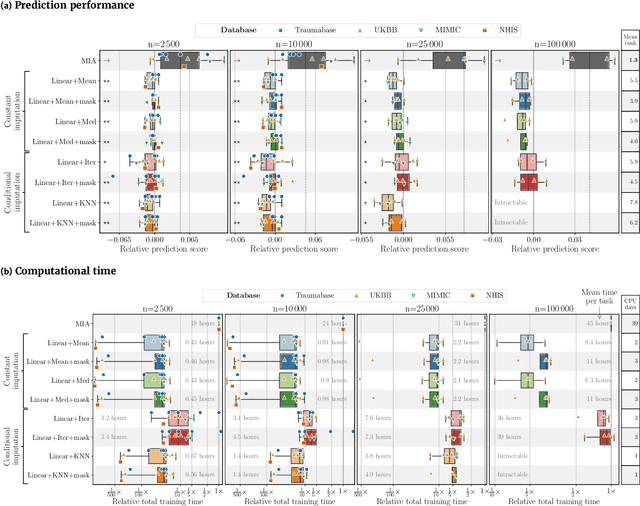
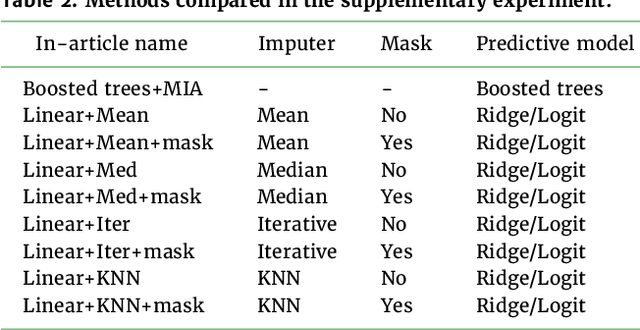
Abstract:BACKGROUND: As databases grow larger, it becomes harder to fully control their collection, and they frequently come with missing values: incomplete observations. These large databases are well suited to train machine-learning models, for instance for forecasting or to extract biomarkers in biomedical settings. Such predictive approaches can use discriminative -- rather than generative -- modeling, and thus open the door to new missing-values strategies. Yet existing empirical evaluations of strategies to handle missing values have focused on inferential statistics. RESULTS: Here we conduct a systematic benchmark of missing-values strategies in predictive models with a focus on large health databases: four electronic health record datasets, a population brain imaging one, a health survey and two intensive care ones. Using gradient-boosted trees, we compare native support for missing values with simple and state-of-the-art imputation prior to learning. We investigate prediction accuracy and computational time. For prediction after imputation, we find that adding an indicator to express which values have been imputed is important, suggesting that the data are missing not at random. Elaborate missing values imputation can improve prediction compared to simple strategies but requires longer computational time on large data. Learning trees that model missing values-with missing incorporated attribute-leads to robust, fast, and well-performing predictive modeling. CONCLUSIONS: Native support for missing values in supervised machine learning predicts better than state-of-the-art imputation with much less computational cost. When using imputation, it is important to add indicator columns expressing which values have been imputed.
What's a good imputation to predict with missing values?
Jun 01, 2021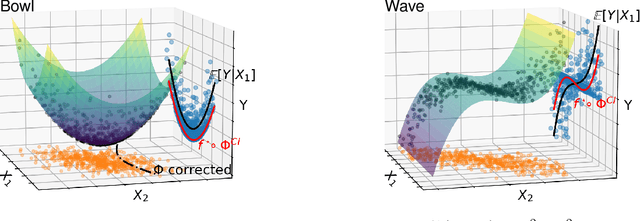
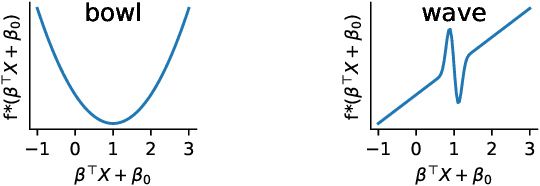
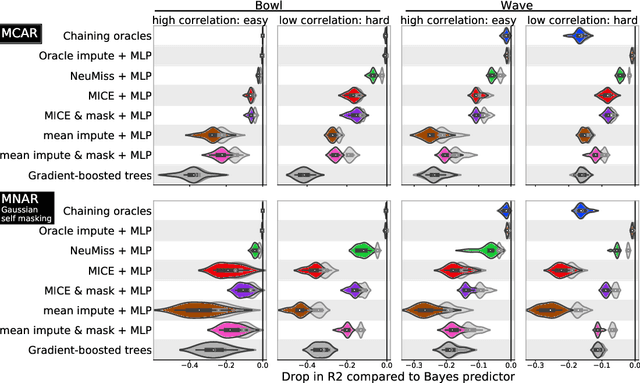
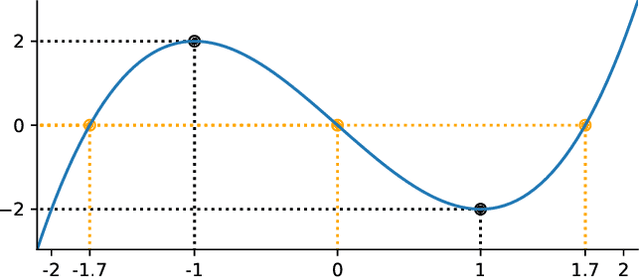
Abstract:How to learn a good predictor on data with missing values? Most efforts focus on first imputing as well as possible and second learning on the completed data to predict the outcome. Yet, this widespread practice has no theoretical grounding. Here we show that for almost all imputation functions, an impute-then-regress procedure with a powerful learner is Bayes optimal. This result holds for all missing-values mechanisms, in contrast with the classic statistical results that require missing-at-random settings to use imputation in probabilistic modeling. Moreover, it implies that perfect conditional imputation may not be needed for good prediction asymptotically. In fact, we show that on perfectly imputed data the best regression function will generally be discontinuous, which makes it hard to learn. Crafting instead the imputation so as to leave the regression function unchanged simply shifts the problem to learning discontinuous imputations. Rather, we suggest that it is easier to learn imputation and regression jointly. We propose such a procedure, adapting NeuMiss, a neural network capturing the conditional links across observed and unobserved variables whatever the missing-value pattern. Experiments confirm that joint imputation and regression through NeuMiss is better than various two step procedures in our experiments with finite number of samples.
Neumann networks: differential programming for supervised learning with missing values
Jul 03, 2020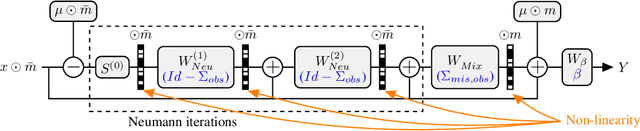
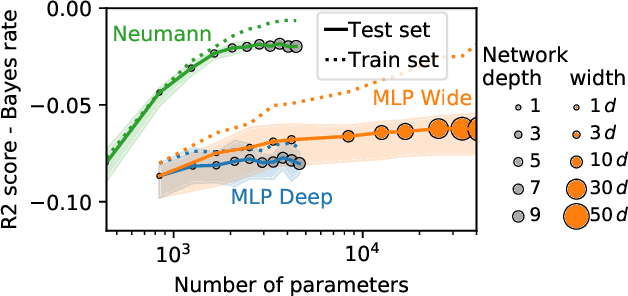
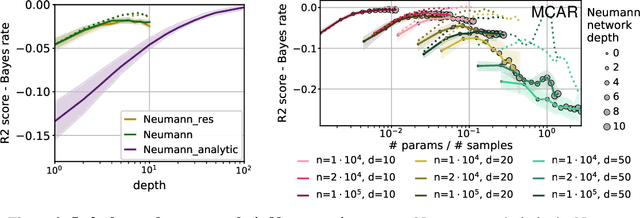
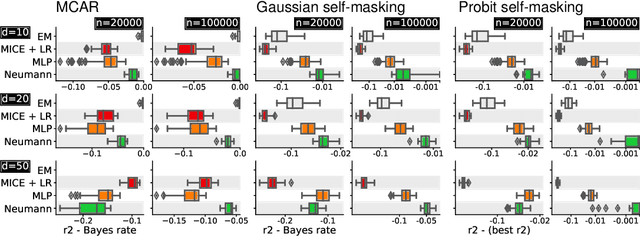
Abstract:The presence of missing values makes supervised learning much more challenging. Indeed, previous work has shown that even when the response is a linear function of the complete data, the optimal predictor is a complex function of the observed entries and the missingness indicator. As a result, the computational or sample complexities of consistent approaches depend on the number of missing patterns, which can be exponential in the number of dimensions. In this work, we derive the analytical form of the optimal predictor under a linearity assumption and various missing data mechanisms including Missing at Random (MAR) and self-masking (Missing Not At Random). Based on a Neumann series approximation of the optimal predictor, we propose a new principled architecture, named Neumann networks. Their originality and strength comes from the use of a new type of non-linearity: the multiplication by the missingness indicator. We provide an upper bound on the Bayes risk of Neumann networks, and show that they have good predictive accuracy with both a number of parameters and a computational complexity independent of the number of missing data patterns. As a result they scale well to problems with many features, and remain statistically efficient for medium-sized samples. Moreover, we show that, contrary to procedures using EM or imputation, they are robust to the missing data mechanism, including difficult MNAR settings such as self-masking.
Linear predictor on linearly-generated data with missing values: non consistency and solutions
Feb 03, 2020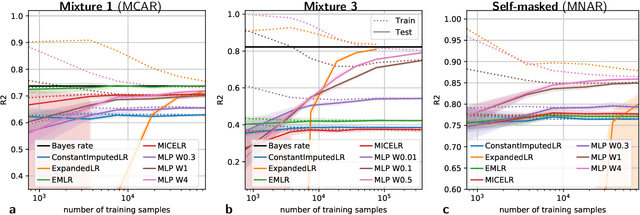
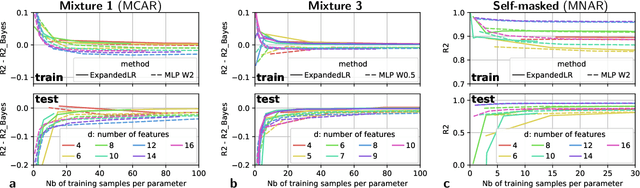
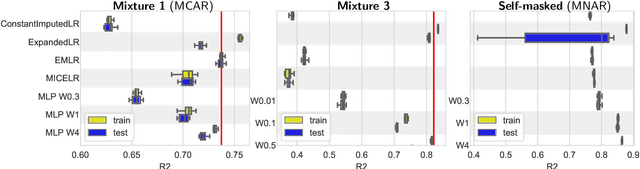
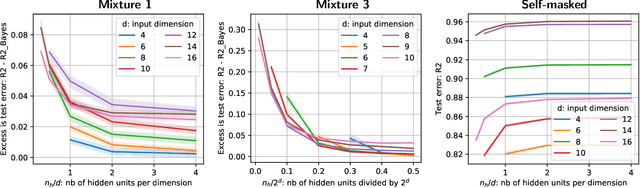
Abstract:We consider building predictors when the data have missing values. We study the seemingly-simple case where the target to predict is a linear function of the fully-observed data and we show that, in the presence of missing values, the optimal predictor may not be linear. In the particular Gaussian case, it can be written as a linear function of multiway interactions between the observed data and the various missing-value indicators. Due to its intrinsic complexity, we study a simple approximation and prove generalization bounds with finite samples, highlighting regimes for which each method performs best. We then show that multilayer perceptrons with ReLU activation functions can be consistent, and can explore good trade-offs between the true model and approximations. Our study highlights the interesting family of models that are beneficial to fit with missing values depending on the amount of data available.
WHInter: A Working set algorithm for High-dimensional sparse second order Interaction models
Feb 16, 2018

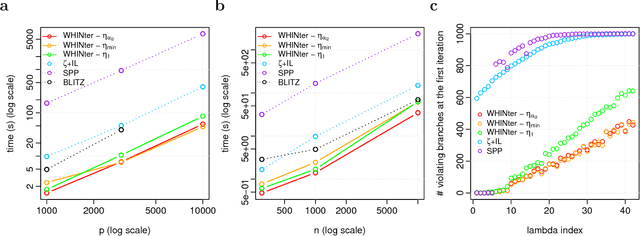
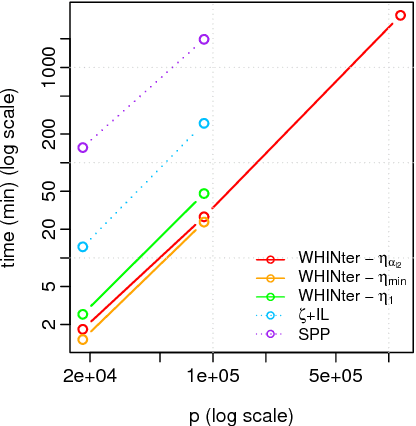
Abstract:Learning sparse linear models with two-way interactions is desirable in many application domains such as genomics. l1-regularised linear models are popular to estimate sparse models, yet standard implementations fail to address specifically the quadratic explosion of candidate two-way interactions in high dimensions, and typically do not scale to genetic data with hundreds of thousands of features. Here we present WHInter, a working set algorithm to solve large l1-regularised problems with two-way interactions for binary design matrices. The novelty of WHInter stems from a new bound to efficiently identify working sets while avoiding to scan all features, and on fast computations inspired from solutions to the maximum inner product search problem. We apply WHInter to simulated and real genetic data and show that it is more scalable and two orders of magnitude faster than the state of the art.
Supervised Quantile Normalisation
Jun 01, 2017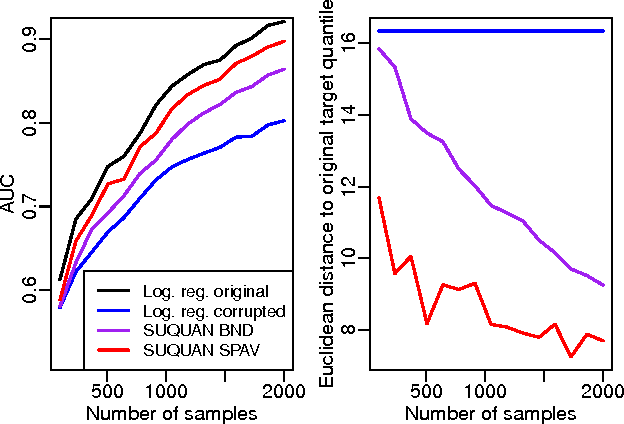

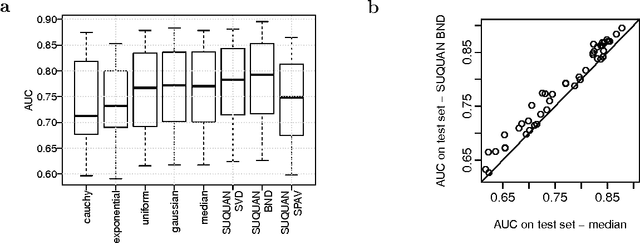
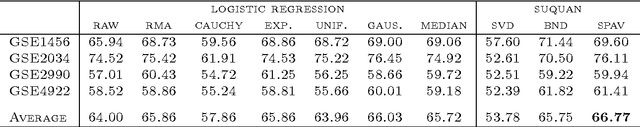
Abstract:Quantile normalisation is a popular normalisation method for data subject to unwanted variations such as images, speech, or genomic data. It applies a monotonic transformation to the feature values of each sample to ensure that after normalisation, they follow the same target distribution for each sample. Choosing a "good" target distribution remains however largely empirical and heuristic, and is usually done independently of the subsequent analysis of normalised data. We propose instead to couple the quantile normalisation step with the subsequent analysis, and to optimise the target distribution jointly with the other parameters in the analysis. We illustrate this principle on the problem of estimating a linear model over normalised data, and show that it leads to a particular low-rank matrix regression problem that can be solved efficiently. We illustrate the potential of our method, which we term SUQUAN, on simulated data, images and genomic data, where it outperforms standard quantile normalisation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge