Supervised Quantile Normalisation
Paper and Code
Jun 01, 2017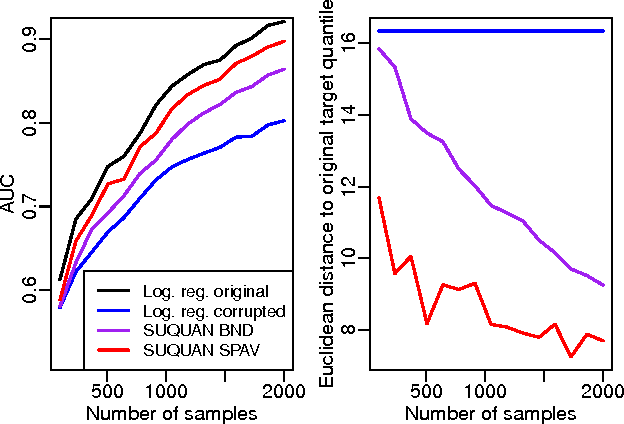

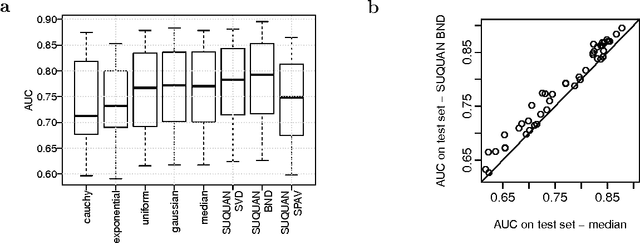
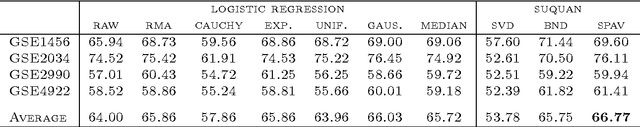
Quantile normalisation is a popular normalisation method for data subject to unwanted variations such as images, speech, or genomic data. It applies a monotonic transformation to the feature values of each sample to ensure that after normalisation, they follow the same target distribution for each sample. Choosing a "good" target distribution remains however largely empirical and heuristic, and is usually done independently of the subsequent analysis of normalised data. We propose instead to couple the quantile normalisation step with the subsequent analysis, and to optimise the target distribution jointly with the other parameters in the analysis. We illustrate this principle on the problem of estimating a linear model over normalised data, and show that it leads to a particular low-rank matrix regression problem that can be solved efficiently. We illustrate the potential of our method, which we term SUQUAN, on simulated data, images and genomic data, where it outperforms standard quantile normalisation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge