WHInter: A Working set algorithm for High-dimensional sparse second order Interaction models
Paper and Code
Feb 16, 2018

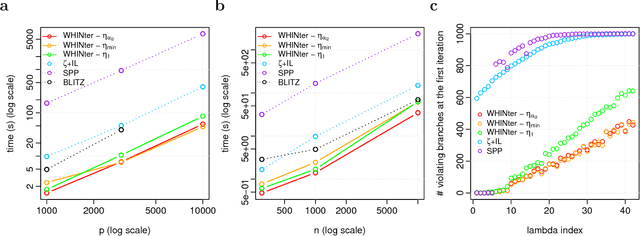
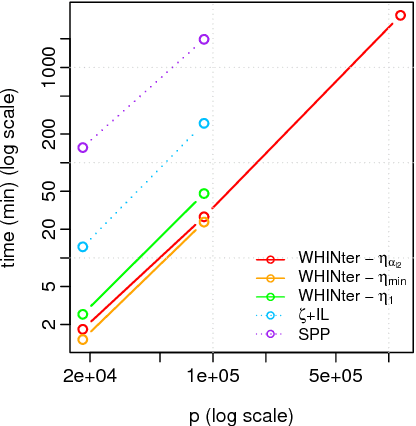
Learning sparse linear models with two-way interactions is desirable in many application domains such as genomics. l1-regularised linear models are popular to estimate sparse models, yet standard implementations fail to address specifically the quadratic explosion of candidate two-way interactions in high dimensions, and typically do not scale to genetic data with hundreds of thousands of features. Here we present WHInter, a working set algorithm to solve large l1-regularised problems with two-way interactions for binary design matrices. The novelty of WHInter stems from a new bound to efficiently identify working sets while avoiding to scan all features, and on fast computations inspired from solutions to the maximum inner product search problem. We apply WHInter to simulated and real genetic data and show that it is more scalable and two orders of magnitude faster than the state of the art.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge