Manan Shah
PyG 2.0: Scalable Learning on Real World Graphs
Jul 22, 2025Abstract:PyG (PyTorch Geometric) has evolved significantly since its initial release, establishing itself as a leading framework for Graph Neural Networks. In this paper, we present Pyg 2.0 (and its subsequent minor versions), a comprehensive update that introduces substantial improvements in scalability and real-world application capabilities. We detail the framework's enhanced architecture, including support for heterogeneous and temporal graphs, scalable feature/graph stores, and various optimizations, enabling researchers and practitioners to tackle large-scale graph learning problems efficiently. Over the recent years, PyG has been supporting graph learning in a large variety of application areas, which we will summarize, while providing a deep dive into the important areas of relational deep learning and large language modeling.
The Convergent Ethics of AI? Analyzing Moral Foundation Priorities in Large Language Models with a Multi-Framework Approach
Apr 27, 2025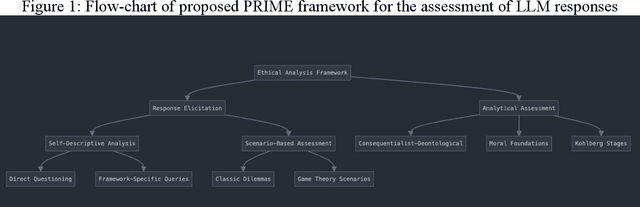
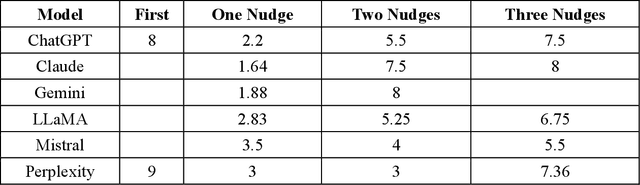
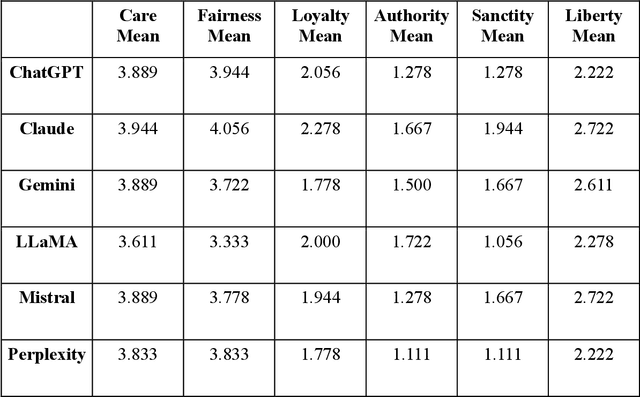
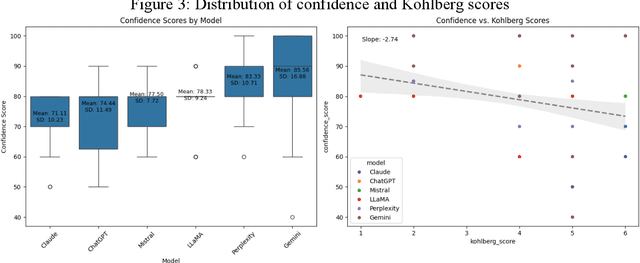
Abstract:As large language models (LLMs) are increasingly deployed in consequential decision-making contexts, systematically assessing their ethical reasoning capabilities becomes a critical imperative. This paper introduces the Priorities in Reasoning and Intrinsic Moral Evaluation (PRIME) framework--a comprehensive methodology for analyzing moral priorities across foundational ethical dimensions including consequentialist-deontological reasoning, moral foundations theory, and Kohlberg's developmental stages. We apply this framework to six leading LLMs through a dual-protocol approach combining direct questioning and response analysis to established ethical dilemmas. Our analysis reveals striking patterns of convergence: all evaluated models demonstrate strong prioritization of care/harm and fairness/cheating foundations while consistently underweighting authority, loyalty, and sanctity dimensions. Through detailed examination of confidence metrics, response reluctance patterns, and reasoning consistency, we establish that contemporary LLMs (1) produce decisive ethical judgments, (2) demonstrate notable cross-model alignment in moral decision-making, and (3) generally correspond with empirically established human moral preferences. This research contributes a scalable, extensible methodology for ethical benchmarking while highlighting both the promising capabilities and systematic limitations in current AI moral reasoning architectures--insights critical for responsible development as these systems assume increasingly significant societal roles.
Auditing the Ethical Logic of Generative AI Models
Apr 24, 2025Abstract:As generative AI models become increasingly integrated into high-stakes domains, the need for robust methods to evaluate their ethical reasoning becomes increasingly important. This paper introduces a five-dimensional audit model -- assessing Analytic Quality, Breadth of Ethical Considerations, Depth of Explanation, Consistency, and Decisiveness -- to evaluate the ethical logic of leading large language models (LLMs). Drawing on traditions from applied ethics and higher-order thinking, we present a multi-battery prompt approach, including novel ethical dilemmas, to probe the models' reasoning across diverse contexts. We benchmark seven major LLMs finding that while models generally converge on ethical decisions, they vary in explanatory rigor and moral prioritization. Chain-of-Thought prompting and reasoning-optimized models significantly enhance performance on our audit metrics. This study introduces a scalable methodology for ethical benchmarking of AI systems and highlights the potential for AI to complement human moral reasoning in complex decision-making contexts.
MirrorVerse: Pushing Diffusion Models to Realistically Reflect the World
Apr 21, 2025Abstract:Diffusion models have become central to various image editing tasks, yet they often fail to fully adhere to physical laws, particularly with effects like shadows, reflections, and occlusions. In this work, we address the challenge of generating photorealistic mirror reflections using diffusion-based generative models. Despite extensive training data, existing diffusion models frequently overlook the nuanced details crucial to authentic mirror reflections. Recent approaches have attempted to resolve this by creating synhetic datasets and framing reflection generation as an inpainting task; however, they struggle to generalize across different object orientations and positions relative to the mirror. Our method overcomes these limitations by introducing key augmentations into the synthetic data pipeline: (1) random object positioning, (2) randomized rotations, and (3) grounding of objects, significantly enhancing generalization across poses and placements. To further address spatial relationships and occlusions in scenes with multiple objects, we implement a strategy to pair objects during dataset generation, resulting in a dataset robust enough to handle these complex scenarios. Achieving generalization to real-world scenes remains a challenge, so we introduce a three-stage training curriculum to develop the MirrorFusion 2.0 model to improve real-world performance. We provide extensive qualitative and quantitative evaluations to support our approach. The project page is available at: https://mirror-verse.github.io/.
Analyzing the Ethical Logic of Six Large Language Models
Jan 15, 2025
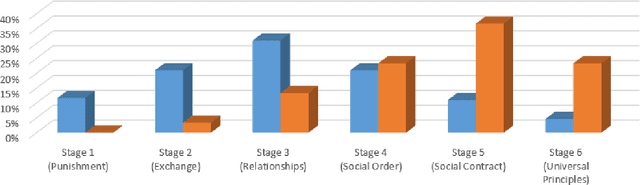

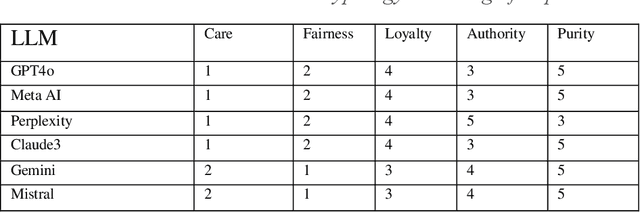
Abstract:This study examines the ethical reasoning of six prominent generative large language models: OpenAI GPT-4o, Meta LLaMA 3.1, Perplexity, Anthropic Claude 3.5 Sonnet, Google Gemini, and Mistral 7B. The research explores how these models articulate and apply ethical logic, particularly in response to moral dilemmas such as the Trolley Problem, and Heinz Dilemma. Departing from traditional alignment studies, the study adopts an explainability-transparency framework, prompting models to explain their ethical reasoning. This approach is analyzed through three established ethical typologies: the consequentialist-deontological analytic, Moral Foundations Theory, and the Kohlberg Stages of Moral Development Model. Findings reveal that LLMs exhibit largely convergent ethical logic, marked by a rationalist, consequentialist emphasis, with decisions often prioritizing harm minimization and fairness. Despite similarities in pre-training and model architecture, a mixture of nuanced and significant differences in ethical reasoning emerge across models, reflecting variations in fine-tuning and post-training processes. The models consistently display erudition, caution, and self-awareness, presenting ethical reasoning akin to a graduate-level discourse in moral philosophy. In striking uniformity these systems all describe their ethical reasoning as more sophisticated than what is characteristic of typical human moral logic.
ContextGNN: Beyond Two-Tower Recommendation Systems
Nov 29, 2024



Abstract:Recommendation systems predominantly utilize two-tower architectures, which evaluate user-item rankings through the inner product of their respective embeddings. However, one key limitation of two-tower models is that they learn a pair-agnostic representation of users and items. In contrast, pair-wise representations either scale poorly due to their quadratic complexity or are too restrictive on the candidate pairs to rank. To address these issues, we introduce Context-based Graph Neural Networks (ContextGNNs), a novel deep learning architecture for link prediction in recommendation systems. The method employs a pair-wise representation technique for familiar items situated within a user's local subgraph, while leveraging two-tower representations to facilitate the recommendation of exploratory items. A final network then predicts how to fuse both pair-wise and two-tower recommendations into a single ranking of items. We demonstrate that ContextGNN is able to adapt to different data characteristics and outperforms existing methods, both traditional and GNN-based, on a diverse set of practical recommendation tasks, improving performance by 20% on average.
Reproducibility Study of CDUL: CLIP-Driven Unsupervised Learning for Multi-Label Image Classification
May 19, 2024



Abstract:This report is a reproducibility study of the paper "CDUL: CLIP-Driven Unsupervised Learning for Multi-Label Image Classification" (Abdelfattah et al, ICCV 2023). Our report makes the following contributions: (1) We provide a reproducible, well commented and open-sourced code implementation for the entire method specified in the original paper. (2) We try to verify the effectiveness of the novel aggregation strategy which uses the CLIP model to initialize the pseudo labels for the subsequent unsupervised multi-label image classification task. (3) We try to verify the effectiveness of the gradient-alignment training method specified in the original paper, which is used to update the network parameters and pseudo labels. The code can be found at https://github.com/cs-mshah/CDUL
Contextual Code Switching for Machine Translation using Language Models
Dec 20, 2023Abstract:Large language models (LLMs) have exerted a considerable impact on diverse language-related tasks in recent years. Their demonstrated state-of-the-art performance is achieved through methodologies such as zero-shot or few-shot prompting. These models undergo training on extensive datasets that encompass segments of the Internet and subsequently undergo fine-tuning tailored to specific tasks. Notably, they exhibit proficiency in tasks such as translation, summarization, question answering, and creative writing, even in the absence of explicit training for those particular tasks. While they have shown substantial improvement in the multilingual tasks their performance in the code switching, especially for machine translation remains relatively uncharted. In this paper, we present an extensive study on the code switching task specifically for the machine translation task comparing multiple LLMs. Our results indicate that despite the LLMs having promising results in the certain tasks, the models with relatively lesser complexity outperform the multilingual large language models in the machine translation task. We posit that the efficacy of multilingual large language models in contextual code switching is constrained by their training methodologies. In contrast, relatively smaller models, when trained and fine-tuned on bespoke datasets, may yield superior results in comparison to the majority of multilingual models.
Carbon Emission Prediction on the World Bank Dataset for Canada
Nov 26, 2022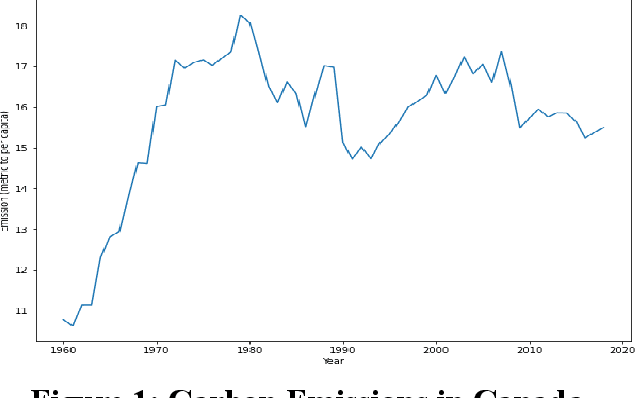
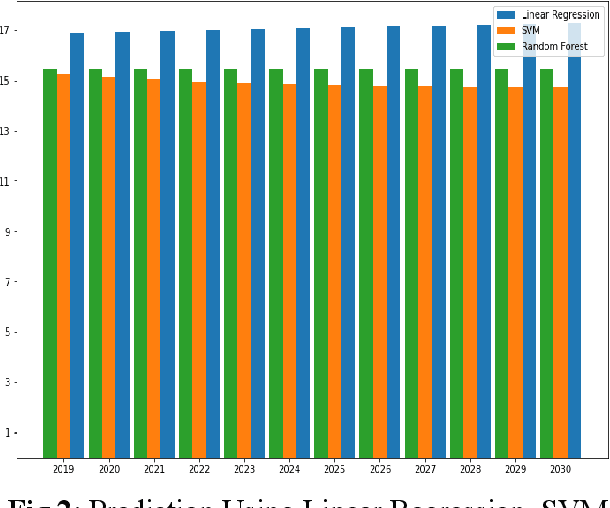
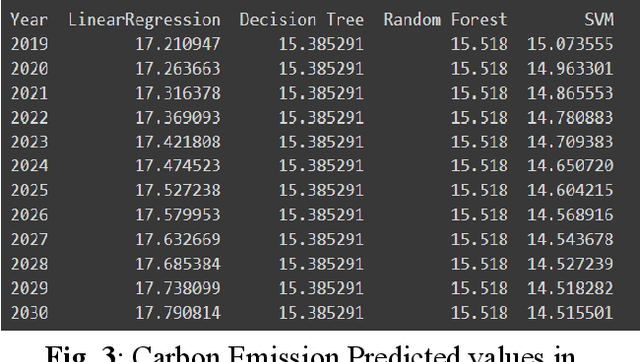
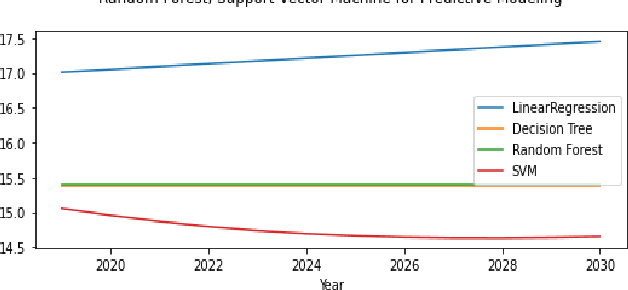
Abstract:The continuous rise in CO2 emission into the environment is one of the most crucial issues facing the whole world. Many countries are making crucial decisions to control their carbon footprints to escape some of their catastrophic outcomes. There has been a lot of research going on to project the amount of carbon emissions in the future, which can help us to develop innovative techniques to deal with it in advance. Machine learning is one of the most advanced and efficient techniques for predicting the amount of carbon emissions from current data. This paper provides the methods for predicting carbon emissions (CO2 emissions) for the next few years. The predictions are based on data from the past 50 years. The dataset, which is used for making the prediction, is collected from World Bank datasets. This dataset contains CO2 emissions (metric tons per capita) of all the countries from 1960 to 2018. Our method consists of using machine learning techniques to take the idea of what carbon emission measures will look like in the next ten years and project them onto the dataset taken from the World Bank's data repository. The purpose of this research is to compare how different machine learning models (Decision Tree, Linear Regression, Random Forest, and Support Vector Machine) perform on a similar dataset and measure the difference between their predictions.
Advancement of Deep Learning in Pneumonia and Covid-19 Classification and Localization: A Qualitative and Quantitative Analysis
Nov 16, 2021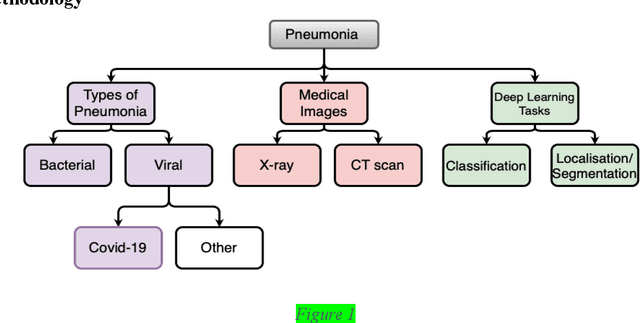
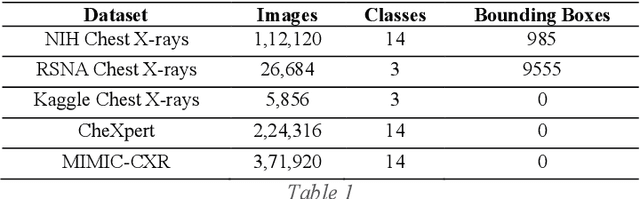
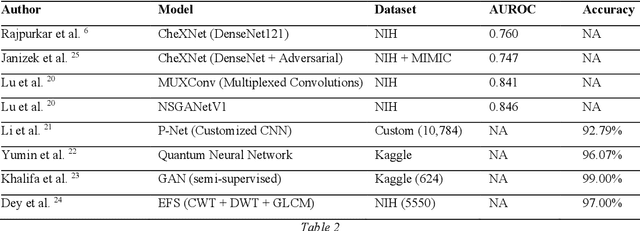
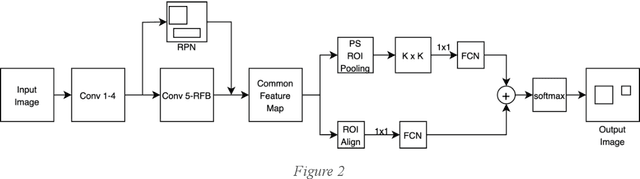
Abstract:Around 450 million people are affected by pneumonia every year which results in 2.5 million deaths. Covid-19 has also affected 181 million people which has lead to 3.92 million casualties. The chances of death in both of these diseases can be significantly reduced if they are diagnosed early. However, the current methods of diagnosing pneumonia (complaints + chest X-ray) and covid-19 (RT-PCR) require the presence of expert radiologists and time, respectively. With the help of Deep Learning models, pneumonia and covid-19 can be detected instantly from Chest X-rays or CT scans. This way, the process of diagnosing Pneumonia/Covid-19 can be made more efficient and widespread. In this paper, we aim to elicit, explain, and evaluate, qualitatively and quantitatively, major advancements in deep learning methods aimed at detecting or localizing community-acquired pneumonia (CAP), viral pneumonia, and covid-19 from images of chest X-rays and CT scans. Being a systematic review, the focus of this paper lies in explaining deep learning model architectures which have either been modified or created from scratch for the task at hand wiwth focus on generalizability. For each model, this paper answers the question of why the model is designed the way it is, the challenges that a particular model overcomes, and the tradeoffs that come with modifying a model to the required specifications. A quantitative analysis of all models described in the paper is also provided to quantify the effectiveness of different models with a similar goal. Some tradeoffs cannot be quantified, and hence they are mentioned explicitly in the qualitative analysis, which is done throughout the paper. By compiling and analyzing a large quantum of research details in one place with all the datasets, model architectures, and results, we aim to provide a one-stop solution to beginners and current researchers interested in this field.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge