Mahdi Imani
MINT: Minimal Information Neuro-Symbolic Tree for Objective-Driven Knowledge-Gap Reasoning and Active Elicitation
Feb 04, 2026Abstract:Joint planning through language-based interactions is a key area of human-AI teaming. Planning problems in the open world often involve various aspects of incomplete information and unknowns, e.g., objects involved, human goals/intents -- thus leading to knowledge gaps in joint planning. We consider the problem of discovering optimal interaction strategies for AI agents to actively elicit human inputs in object-driven planning. To this end, we propose Minimal Information Neuro-Symbolic Tree (MINT) to reason about the impact of knowledge gaps and leverage self-play with MINT to optimize the AI agent's elicitation strategies and queries. More precisely, MINT builds a symbolic tree by making propositions of possible human-AI interactions and by consulting a neural planning policy to estimate the uncertainty in planning outcomes caused by remaining knowledge gaps. Finally, we leverage LLM to search and summarize MINT's reasoning process and curate a set of queries to optimally elicit human inputs for best planning performance. By considering a family of extended Markov decision processes with knowledge gaps, we analyze the return guarantee for a given MINT with active human elicitation. Our evaluation on three benchmarks involving unseen/unknown objects of increasing realism shows that MINT-based planning attains near-expert returns by issuing a limited number of questions per task while achieving significantly improved rewards and success rates.
Manifold-Constrained Energy-Based Transition Models for Offline Reinforcement Learning
Feb 02, 2026Abstract:Model-based offline reinforcement learning is brittle under distribution shift: policy improvement drives rollouts into state--action regions weakly supported by the dataset, where compounding model error yields severe value overestimation. We propose Manifold-Constrained Energy-based Transition Models (MC-ETM), which train conditional energy-based transition models using a manifold projection--diffusion negative sampler. MC-ETM learns a latent manifold of next states and generates near-manifold hard negatives by perturbing latent codes and running Langevin dynamics in latent space with the learned conditional energy, sharpening the energy landscape around the dataset support and improving sensitivity to subtle out-of-distribution deviations. For policy optimization, the learned energy provides a single reliability signal: rollouts are truncated when the minimum energy over sampled next states exceeds a threshold, and Bellman backups are stabilized via pessimistic penalties based on Q-value-level dispersion across energy-guided samples. We formalize MC-ETM through a hybrid pessimistic MDP formulation and derive a conservative performance bound separating in-support evaluation error from truncation risk. Empirically, MC-ETM improves multi-step dynamics fidelity and yields higher normalized returns on standard offline control benchmarks, particularly under irregular dynamics and sparse data coverage.
Geometry of Drifting MDPs with Path-Integral Stability Certificates
Jan 29, 2026Abstract:Real-world reinforcement learning is often \emph{nonstationary}: rewards and dynamics drift, accelerate, oscillate, and trigger abrupt switches in the optimal action. Existing theory often represents nonstationarity with coarse-scale models that measure \emph{how much} the environment changes, not \emph{how} it changes locally -- even though acceleration and near-ties drive tracking error and policy chattering. We take a geometric view of nonstationary discounted Markov Decision Processes (MDPs) by modeling the environment as a differentiable homotopy path and tracking the induced motion of the optimal Bellman fixed point. This yields a length-curvature-kink signature of intrinsic complexity: cumulative drift, acceleration/oscillation, and action-gap-induced nonsmoothness. We prove a solver-agnostic path-integral stability bound and derive gap-safe feasible regions that certify local stability away from switch regimes. Building on these results, we introduce \textit{Homotopy-Tracking RL (HT-RL)} and \textit{HT-MCTS}, lightweight wrappers that estimate replay-based proxies of length, curvature, and near-tie proximity online and adapt learning or planning intensity accordingly. Experiments show improved tracking and dynamic regret over matched static baselines, with the largest gains in oscillatory and switch-prone regimes.
ACDZero: Graph-Embedding-Based Tree Search for Mastering Automated Cyber Defense
Jan 05, 2026Abstract:Automated cyber defense (ACD) seeks to protect computer networks with minimal or no human intervention, reacting to intrusions by taking corrective actions such as isolating hosts, resetting services, deploying decoys, or updating access controls. However, existing approaches for ACD, such as deep reinforcement learning (RL), often face difficult exploration in complex networks with large decision/state spaces and thus require an expensive amount of samples. Inspired by the need to learn sample-efficient defense policies, we frame ACD in CAGE Challenge 4 (CAGE-4 / CC4) as a context-based partially observable Markov decision problem and propose a planning-centric defense policy based on Monte Carlo Tree Search (MCTS). It explicitly models the exploration-exploitation tradeoff in ACD and uses statistical sampling to guide exploration and decision making. We make novel use of graph neural networks (GNNs) to embed observations from the network as attributed graphs, to enable permutation-invariant reasoning over hosts and their relationships. To make our solution practical in complex search spaces, we guide MCTS with learned graph embeddings and priors over graph-edit actions, combining model-free generalization and policy distillation with look-ahead planning. We evaluate the resulting agent on CC4 scenarios involving diverse network structures and adversary behaviors, and show that our search-guided, graph-embedding-based planning improves defense reward and robustness relative to state-of-the-art RL baselines.
Draft and Refine with Visual Experts
Nov 14, 2025Abstract:While recent Large Vision-Language Models (LVLMs) exhibit strong multimodal reasoning abilities, they often produce ungrounded or hallucinated responses because they rely too heavily on linguistic priors instead of visual evidence. This limitation highlights the absence of a quantitative measure of how much these models actually use visual information during reasoning. We propose Draft and Refine (DnR), an agent framework driven by a question-conditioned utilization metric. The metric quantifies the model's reliance on visual evidence by first constructing a query-conditioned relevance map to localize question-specific cues and then measuring dependence through relevance-guided probabilistic masking. Guided by this metric, the DnR agent refines its initial draft using targeted feedback from external visual experts. Each expert's output (such as boxes or masks) is rendered as visual cues on the image, and the model is re-queried to select the response that yields the largest improvement in utilization. This process strengthens visual grounding without retraining or architectural changes. Experiments across VQA and captioning benchmarks show consistent accuracy gains and reduced hallucination, demonstrating that measuring visual utilization provides a principled path toward more interpretable and evidence-driven multimodal agent systems.
Global Optimization on Graph-Structured Data via Gaussian Processes with Spectral Representations
Nov 11, 2025Abstract:Bayesian optimization (BO) is a powerful framework for optimizing expensive black-box objectives, yet extending it to graph-structured domains remains challenging due to the discrete and combinatorial nature of graphs. Existing approaches often rely on either full graph topology-impractical for large or partially observed graphs-or incremental exploration, which can lead to slow convergence. We introduce a scalable framework for global optimization over graphs that employs low-rank spectral representations to build Gaussian process (GP) surrogates from sparse structural observations. The method jointly infers graph structure and node representations through learnable embeddings, enabling efficient global search and principled uncertainty estimation even with limited data. We also provide theoretical analysis establishing conditions for accurate recovery of underlying graph structure under different sampling regimes. Experiments on synthetic and real-world datasets demonstrate that our approach achieves faster convergence and improved optimization performance compared to prior methods.
Collaborative AI Teaming in Unknown Environments via Active Goal Deduction
Mar 22, 2024Abstract:With the advancements of artificial intelligence (AI), we're seeing more scenarios that require AI to work closely with other agents, whose goals and strategies might not be known beforehand. However, existing approaches for training collaborative agents often require defined and known reward signals and cannot address the problem of teaming with unknown agents that often have latent objectives/rewards. In response to this challenge, we propose teaming with unknown agents framework, which leverages kernel density Bayesian inverse learning method for active goal deduction and utilizes pre-trained, goal-conditioned policies to enable zero-shot policy adaptation. We prove that unbiased reward estimates in our framework are sufficient for optimal teaming with unknown agents. We further evaluate the framework of redesigned multi-agent particle and StarCraft II micromanagement environments with diverse unknown agents of different behaviors/rewards. Empirical results demonstrate that our framework significantly advances the teaming performance of AI and unknown agents in a wide range of collaborative scenarios.
Bayesian Optimization through Gaussian Cox Process Models for Spatio-temporal Data
Jan 25, 2024Abstract:Bayesian optimization (BO) has established itself as a leading strategy for efficiently optimizing expensive-to-evaluate functions. Existing BO methods mostly rely on Gaussian process (GP) surrogate models and are not applicable to (doubly-stochastic) Gaussian Cox processes, where the observation process is modulated by a latent intensity function modeled as a GP. In this paper, we propose a novel maximum a posteriori inference of Gaussian Cox processes. It leverages the Laplace approximation and change of kernel technique to transform the problem into a new reproducing kernel Hilbert space, where it becomes more tractable computationally. It enables us to obtain both a functional posterior of the latent intensity function and the covariance of the posterior, thus extending existing works that often focus on specific link functions or estimating the posterior mean. Using the result, we propose a BO framework based on the Gaussian Cox process model and further develop a Nystr\"om approximation for efficient computation. Extensive evaluations on various synthetic and real-world datasets demonstrate significant improvement over state-of-the-art inference solutions for Gaussian Cox processes, as well as effective BO with a wide range of acquisition functions designed through the underlying Gaussian Cox process model.
A Bayesian Optimization Framework for Finding Local Optima in Expensive Multi-Modal Functions
Oct 13, 2022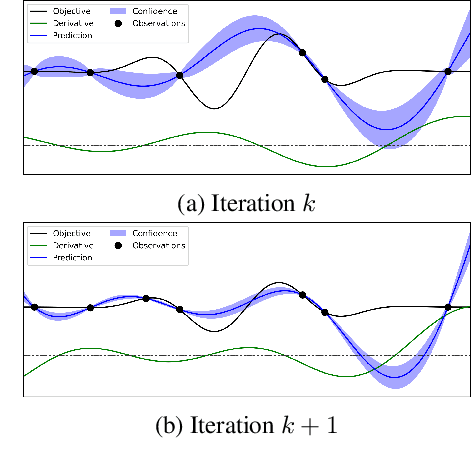
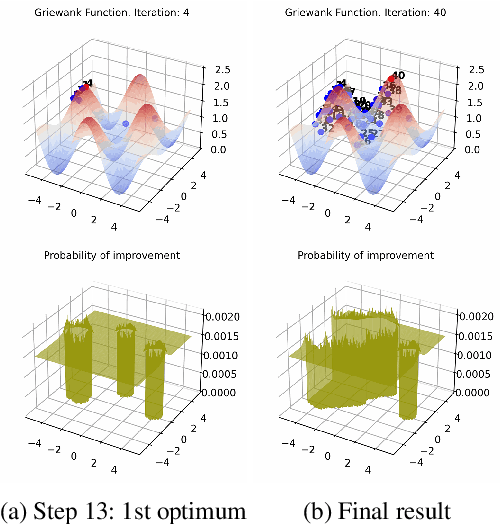
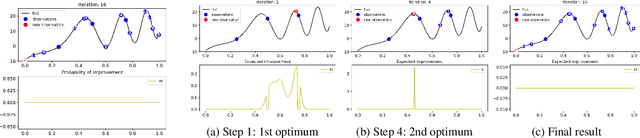
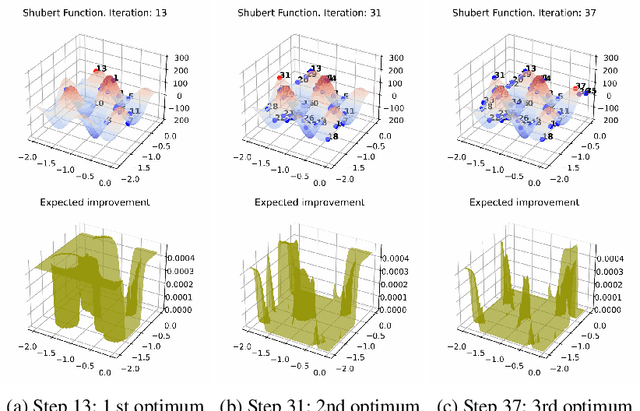
Abstract:Bayesian optimization (BO) is a popular global optimization scheme for sample-efficient optimization in domains with expensive function evaluations. The existing BO techniques are capable of finding a single global optimum solution. However, finding a set of global and local optimum solutions is crucial in a wide range of real-world problems, as implementing some of the optimal solutions might not be feasible due to various practical restrictions (e.g., resource limitation, physical constraints, etc.). In such domains, if multiple solutions are known, the implementation can be quickly switched to another solution, and the best possible system performance can still be obtained. This paper develops a multi-modal BO framework to effectively find a set of local/global solutions for expensive-to-evaluate multi-modal objective functions. We consider the standard BO setting with Gaussian process regression representing the objective function. We analytically derive the joint distribution of the objective function and its first-order gradients. This joint distribution is used in the body of the BO acquisition functions to search for local optima during the optimization process. We introduce variants of the well-known BO acquisition functions to the multi-modal setting and demonstrate the performance of the proposed framework in locating a set of local optimum solutions using multiple optimization problems.
Inference of Regulatory Networks Through Temporally Sparse Data
Jul 21, 2022


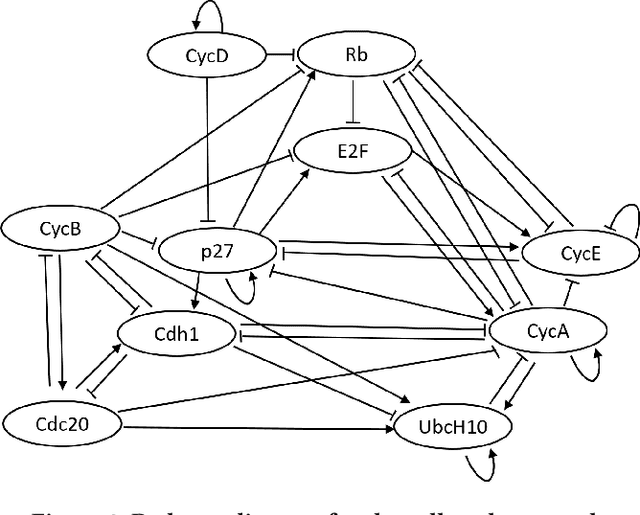
Abstract:A major goal in genomics is to properly capture the complex dynamical behaviors of gene regulatory networks (GRNs). This includes inferring the complex interactions between genes, which can be used for a wide range of genomics analyses, including diagnosis or prognosis of diseases and finding effective treatments for chronic diseases such as cancer. Boolean networks have emerged as a successful class of models for capturing the behavior of GRNs. In most practical settings, inference of GRNs should be achieved through limited and temporally sparse genomics data. A large number of genes in GRNs leads to a large possible topology candidate space, which often cannot be exhaustively searched due to the limitation in computational resources. This paper develops a scalable and efficient topology inference for GRNs using Bayesian optimization and kernel-based methods. Rather than an exhaustive search over possible topologies, the proposed method constructs a Gaussian Process (GP) with a topology-inspired kernel function to account for correlation in the likelihood function. Then, using the posterior distribution of the GP model, the Bayesian optimization efficiently searches for the topology with the highest likelihood value by optimally balancing between exploration and exploitation. The performance of the proposed method is demonstrated through comprehensive numerical experiments using a well-known mammalian cell-cycle network.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge