Madhu Reddiboina
Enhancing Generalization in Audio Deepfake Detection: A Neural Collapse based Sampling and Training Approach
Apr 19, 2024
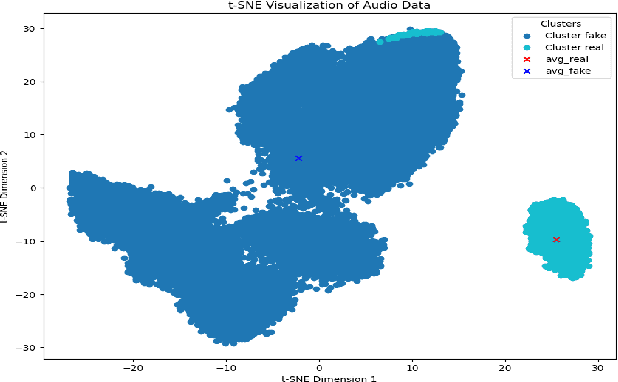

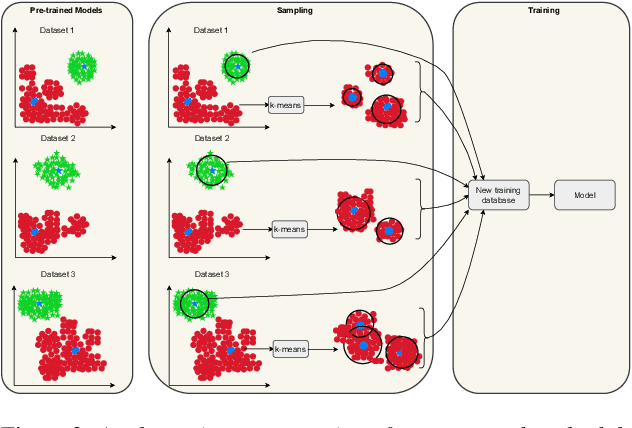
Abstract:Generalization in audio deepfake detection presents a significant challenge, with models trained on specific datasets often struggling to detect deepfakes generated under varying conditions and unknown algorithms. While collectively training a model using diverse datasets can enhance its generalization ability, it comes with high computational costs. To address this, we propose a neural collapse-based sampling approach applied to pre-trained models trained on distinct datasets to create a new training database. Using ASVspoof 2019 dataset as a proof-of-concept, we implement pre-trained models with Resnet and ConvNext architectures. Our approach demonstrates comparable generalization on unseen data while being computationally efficient, requiring less training data. Evaluation is conducted using the In-the-wild dataset.
Towards the Development of a Real-Time Deepfake Audio Detection System in Communication Platforms
Mar 18, 2024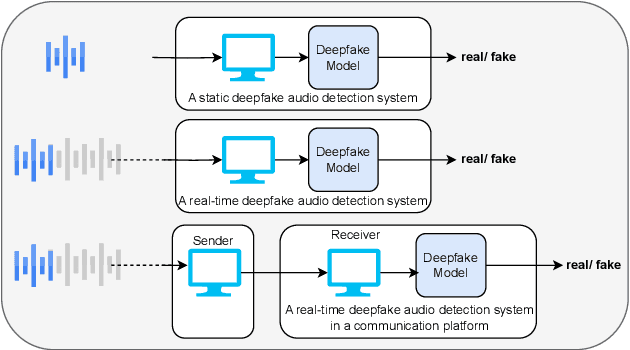
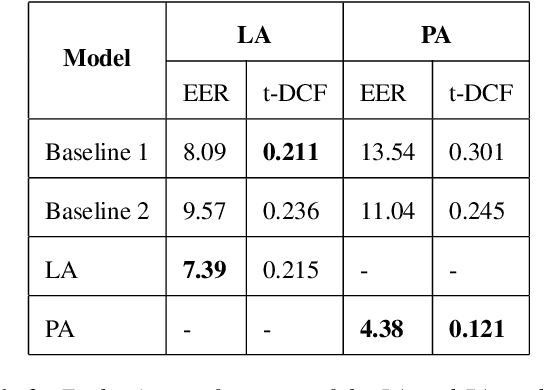
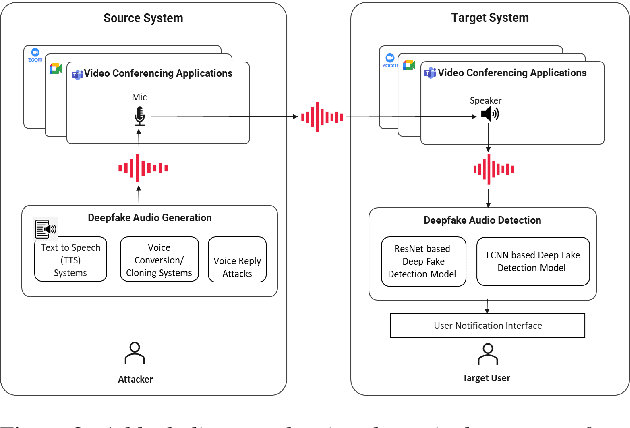
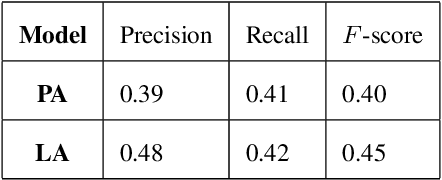
Abstract:Deepfake audio poses a rising threat in communication platforms, necessitating real-time detection for audio stream integrity. Unlike traditional non-real-time approaches, this study assesses the viability of employing static deepfake audio detection models in real-time communication platforms. An executable software is developed for cross-platform compatibility, enabling real-time execution. Two deepfake audio detection models based on Resnet and LCNN architectures are implemented using the ASVspoof 2019 dataset, achieving benchmark performances compared to ASVspoof 2019 challenge baselines. The study proposes strategies and frameworks for enhancing these models, paving the way for real-time deepfake audio detection in communication platforms. This work contributes to the advancement of audio stream security, ensuring robust detection capabilities in dynamic, real-time communication scenarios.
All Data Inclusive, Deep Learning Models to Predict Critical Events in the Medical Information Mart for Intensive Care III Database
Sep 02, 2020Abstract:Intensive care clinicians need reliable clinical practice tools to preempt unexpected critical events that might harm their patients in intensive care units (ICU), to pre-plan timely interventions, and to keep the patient's family well informed. The conventional statistical models are built by curating only a limited number of key variables, which means a vast unknown amount of potentially precious data remains unused. Deep learning models (DLMs) can be leveraged to learn from large complex datasets and construct predictive clinical tools. This retrospective study was performed using 42,818 hospital admissions involving 35,348 patients, which is a subset of the MIMIC-III dataset. Natural language processing (NLP) techniques were applied to build DLMs to predict in-hospital mortality (IHM) and length of stay >=7 days (LOS). Over 75 million events across multiple data sources were processed, resulting in over 355 million tokens. DLMs for predicting IHM using data from all sources (AS) and chart data (CS) achieved an AUC-ROC of 0.9178 and 0.9029, respectively, and PR-AUC of 0.6251 and 0.5701, respectively. DLMs for predicting LOS using AS and CS achieved an AUC-ROC of 0.8806 and 0.8642, respectively, and PR-AUC of 0.6821 and 0.6575, respectively. The observed AUC-ROC difference between models was found to be significant for both IHM and LOS at p=0.05. The observed PR-AUC difference between the models was found to be significant for IHM and statistically insignificant for LOS at p=0.05. In this study, deep learning models were constructed using data combined from a variety of sources in Electronic Health Records (EHRs) such as chart data, input and output events, laboratory values, microbiology events, procedures, notes, and prescriptions. It is possible to predict in-hospital mortality with much better confidence and higher reliability from models built using all sources of data.
2018 Robotic Scene Segmentation Challenge
Feb 03, 2020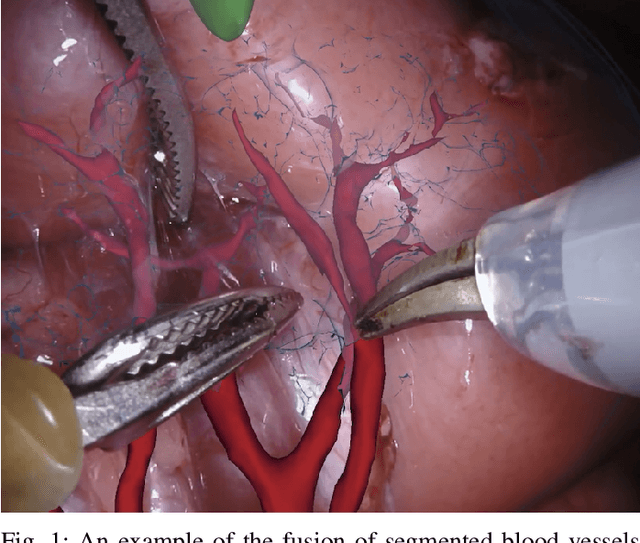
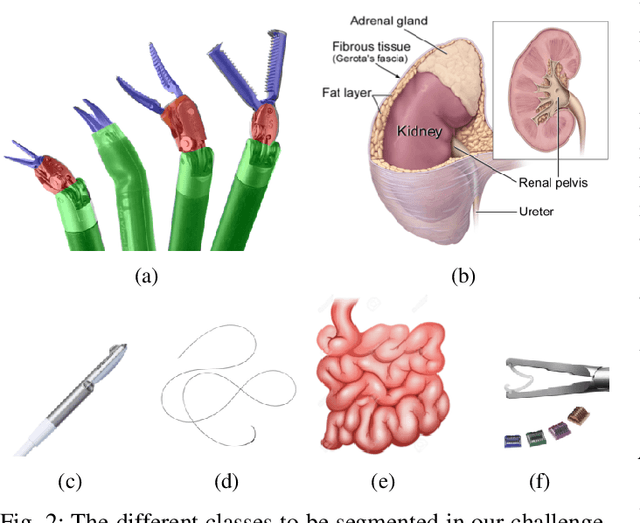
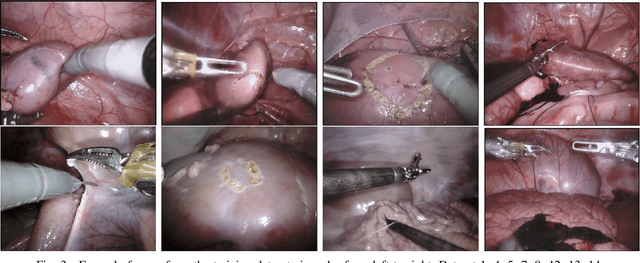
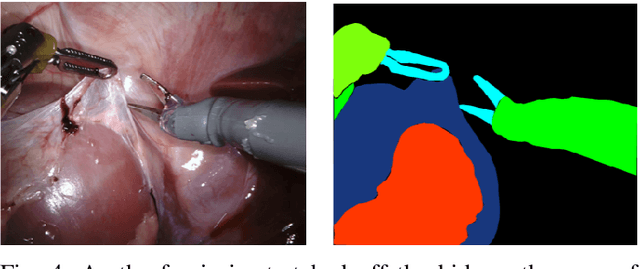
Abstract:In 2015 we began a sub-challenge at the EndoVis workshop at MICCAI in Munich using endoscope images of ex-vivo tissue with automatically generated annotations from robot forward kinematics and instrument CAD models. However, the limited background variation and simple motion rendered the dataset uninformative in learning about which techniques would be suitable for segmentation in real surgery. In 2017, at the same workshop in Quebec we introduced the robotic instrument segmentation dataset with 10 teams participating in the challenge to perform binary, articulating parts and type segmentation of da Vinci instruments. This challenge included realistic instrument motion and more complex porcine tissue as background and was widely addressed with modifications on U-Nets and other popular CNN architectures. In 2018 we added to the complexity by introducing a set of anatomical objects and medical devices to the segmented classes. To avoid over-complicating the challenge, we continued with porcine data which is dramatically simpler than human tissue due to the lack of fatty tissue occluding many organs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge